This page is part of the Genetic Reporting Implementation Guide (v1.1.0: STU 2 Ballot 1) based on FHIR R4. The current version which supercedes this version is 2.0.0. For a full list of available versions, see the Directory of published versions 
This section deals with reporting observations related to cancer genomics. This includes reporting variants in a patient identified as having a somatic source class and reporting germ-line variants with known links to somatic events such as increasing risk for developing particular cancers. Beyond reporting variants, we also provide two “implication” profiles to communicate observed links between variants and relevant knowledge bases such as professional guidelines and publications. This portion of the implementation guide relies on the content in the General Genomic Reporting and Variant Reporting portions of this implementation guide. Somatic genetic reports supplement this information with a somatics-specific implication profile. Implementers of this portion of the implementation guide may also be interested in the Pharmacogenomic Reporting section which deals more generally with medication implications, such as general efficacy, metabolism and increased risks based on patient genetic characteristics.
Some of the somatic profiles make reference to medications and other therapies. Because this implementation guide is intended for international use, it does not mandate the use of any particular code systems for medications or other therapies. Implementations should use the code systems most typically used in their jurisdictions or that are mandated by national FHIR profiles.
While not specifically profiled in this version of the IG, some additional constraints will typically apply to somatic profiles. Patient will typically be mandatory as somatics is not relevant for environmental samples. Body structure will often be identified to allow associating results with specific tumors or lesions - even if multiple samples are drawn over time.
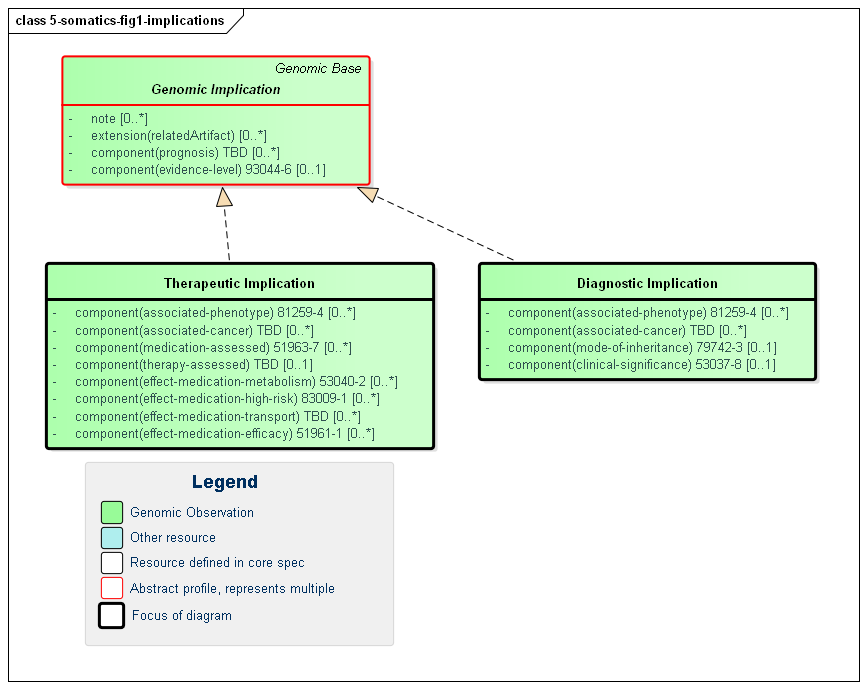
Figure 1: Implication Profiles
(Profile links: Genomic Implication (abstract), Diagnostic Implication Therapeutic Implication)
This implementation guide defines two implication profiles that can both be used for reporting somatic use cases. They each carry a collection of components that provide context for the implication, which can be captured in `Observation.note` as the issuing lab desires.
Diagnostic Implication indicates an association between the identified variant (linked via Observation.derivedFrom) and a particular type of cancer in the associated-phenotype component. The evidence-level and clinical-significance components can convey the evidence-based categorization of the variant for the given cancer type per the AMP/ASCO/CAP guidelines. The prognosis component can also be populated to convey a general indication on the overall outcome for the cancer patient, as described in a linked relatedArtifact.
Therapeutic Implication indicates an association between the identified variant (linked via Observation.derivedFrom) and the efficacy of a particular medication or therapy for the specified cancer. Because this implementation guide is intended for international use, it does not mandate the use of any particular code systems for medications, therapies, or cancer descriptions. Implementations should use the code systems most typically used in their jurisdictions or that are mandated by national FHIR profiles.
For a description of use cases populating other optional components on the above implication profiles, see the Pharmacogenomic Reporting section.