This page is part of the Genetic Reporting Implementation Guide (v1.1.0: STU 2 Ballot 1) based on FHIR R4. The current version which supercedes this version is 2.0.0. For a full list of available versions, see the Directory of published versions 
This section provides guidance on reporting genetic tests that involve sequencing the DNA, RNA or amino acid chains of the specimen. This includes direct sequencing, shotgun sequencing, array-based variant testing and other mechanisms.
Currently, there is one profile used to model most variant information. In future versions of this implementation guide, HL7 may subdivide Variant into multiple sub-profiles with more specific purpose.
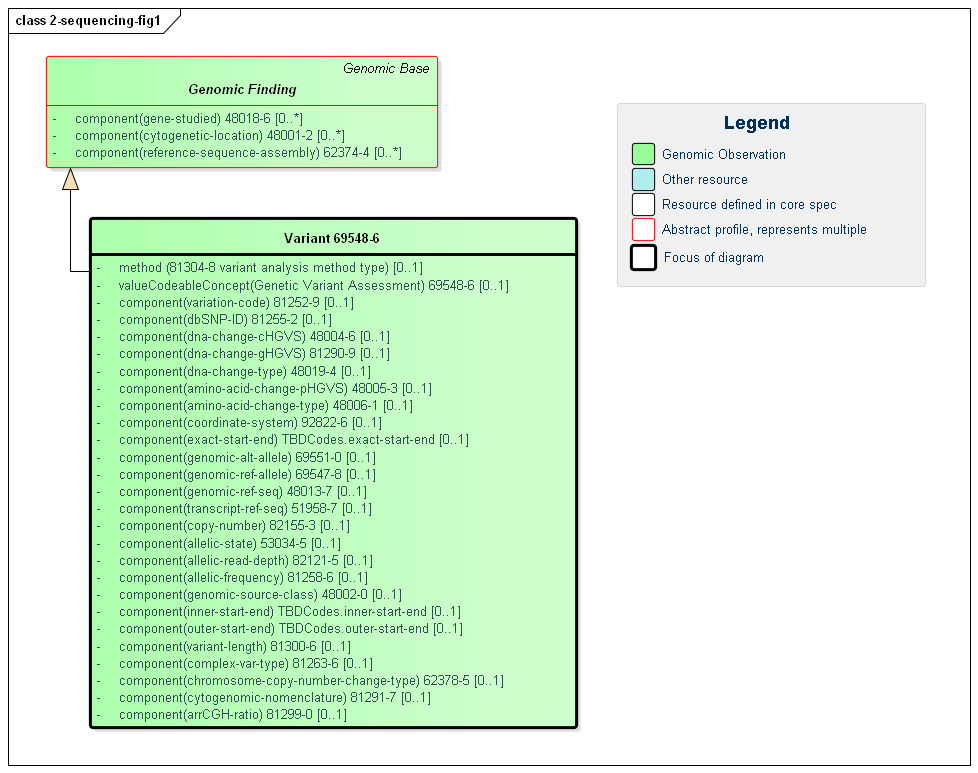
Figure 1: Sequencing Profiles
(Profile links: Variant )
The core of the typical report is a list of identified variants – each with many possible attributes. The variant profile allows a full description of the variant found using properties from a variety of testing approaches and allowing for a variety of descriptive mechanisms. An individual variant may be defined and described by multiple attributes delivered as component values. Most of these components are optional, but labs are encouraged to populate what properties they know. The underlying clinical concept behind each component is mapped to a specific LOINC code. Clinicians are accustomed to receiving descriptive information such as the HGVS genomic and amino acid expression, the type of variant (e.g. deletion) and the reference sequence. The identifiers to online databases can be easily consumed by CDS systems.
Note that there are three coordinate systems one can use when referencing positions within a molecular sequence. For a detailed description of the three options, see here. For clarity and consistency, one must take care not to mix coordinate systems within the same resource instance.
The Variant profile can reference other Variant observations via hasMember when there is a need to represent a named grouping of variants that has a specified clinical effect or phenotype but is not a recognized Haplotype or Genotype. An example would be a compound heterozygote as described here, or other types of findings as in the Complex Variant Type component which uses this LOINC code.