This page is part of the Genetic Reporting Implementation Guide (v1.1.0: STU 2 Ballot 1) based on FHIR R4. The current version which supercedes this version is 2.0.0. For a full list of available versions, see the Directory of published versions 
| Defining URL: | http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/therapeutic-implication |
| Version: | 1.1.0 |
| Name: | TherapeuticImplication |
| Title: | Therapeutic Implication |
| Status: | Active as of 2021-04-13T19:13:37+00:00 |
| Definition: | Profile with properties for observations that convey the potential impact of genomic characteristics on a medication or non-medicinal therapy. |
| Publisher: | HL7 International Clinical Genomics Work Group |
| Copyright: | This material contains content from LOINC (http://loinc.org). LOINC is copyright © 1995-2020, Regenstrief Institute, Inc. and the Logical Observation Identifiers Names and Codes (LOINC) Committee and is available at no cost under the license at http://loinc.org/license. LOINC® is a registered United States trademark of Regenstrief Institute, Inc. |
| Source Resource: | XML / JSON / Turtle |
The official URL for this profile is:
http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/therapeutic-implication
Contents:
The content in this section may be significantly revised prior to the next ballot after initial use and feedback.
This section provides guidance for genomic reporting about the implication of a patient's genetics on the behavior of one or more medications. This includes making recommendations for medication adjustments. This portion of the implementation guide relies on the content in the General Genomic Reporting and Variant Reporting portions of this implementation guide. Pharmacogenomic reports supplement this information with a set of pharmacogenomic-specific implication profiles. Implementers of pharmacogenomic reporting may also be interested in the Somatic Reporting section of this implementation guide.
In general, treatments and medications have a set of indications for use which are not explicitly stated. However, if a pharmacogenomics statement has a specific context of a disease (indication) or a tumor indication use the components "associated phenotype" or "associated cancer" respectively.
To explicitly link medication or treatment implication to oncology, for example, populate the associated-cancer component. E.g. If the associated-cancer component is populated, then effect-medication-efficacy pertains to the medication's efficacy on treating the cancer. For oncology related reporting in general see Somatic Reporting.
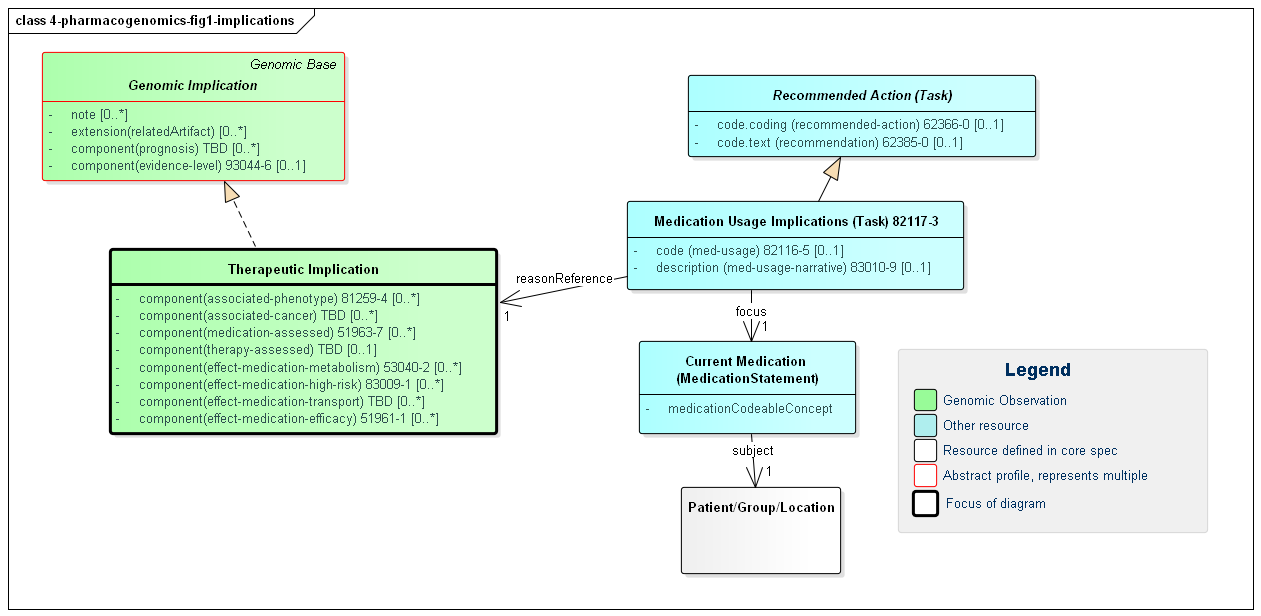
Figure 1: Pharmacogenomic Implications
(Profile links: Genetic Implication, Therapeutic Implication, Medication Usage Implication )
All pharmacogenomic implications are intended to be communicated using components within Therapeutic implication profile. The profile component medication-assessed is the place to communicate the medication whose implication is being described, including a place for the coded value of the medication. The Therapy-assessed component is not intended for medications. Because this is an international profile, no guidance is provided on drug coding systems. The typical or FHIR-mandated jurisdictional code system(s) should be used.
There are several types of medication implication component defined in the profile for use in pharamcogenomics:
To associate a Therapeutic-implication instance with a recommended action (e.g. discontinuing a medication, altering dosage, "choose alternative medication") use Medication Usage Implication, Recommended action, and Current Medication. Use "related artifact" in Therapeutic-implication for supporting documents such as CPIC guidelines.
Medication Usage Implications (task-med-chg) link to therapeutic implications through "reasonReference"and are included in the report as links for RecommendedAction, DiagnosticReport.extension:RecommendedAction. The therapeutic implications do not have a link to medication usage implications (task-med-chg): so rev_include is currently necessary to find the therapeutic implication statements relevant to the medication usage implications.
Note: if multiple medications are used in medication assessed, the usage implication should describe the combined effect. Note 2: Implementers may also be interested in the Somatic Reporting and Pharmacogenomics Reporting sections of this implementation guide.
Description of Profiles, Differentials, Snapshots and how the different presentations work.
This structure is derived from GenomicImplication
This structure is derived from GenomicImplication
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | GenomicImplication | Measurements and simple assertions | |
  | ||||
   | ||||
    | 0..1 | uri | Identity of the terminology system Required Pattern: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
    | 0..1 | code | Symbol in syntax defined by the system Required Pattern: therapeutic-implication | |
  | 0..* | BackboneElement | Associated phenotype | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81259-4 | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..* | BackboneElement | Associated cancer | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-cancer | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..* | BackboneElement | Medication Assessed | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51963-7 | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..* | BackboneElement | Associated cancer | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-therapy | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..1 | BackboneElement | Effect medication metabolism | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53040-2 | |
   | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL3856-3 (preferred) | |
  | 0..1 | BackboneElement | Effect medication high risk | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 83009-1 | |
   | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL2353-2 (extensible) | |
  | 0..* | BackboneElement | Effect medication high risk | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51961-1 | |
   | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL539-8 (preferred) | |
 Documentation for this format Documentation for this format | ||||
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | I | 0..* | GenomicImplication | Measurements and simple assertions |
  | Σ | 0..1 | string | Logical id of this artifact |
  | Σ | 0..1 | Meta | Metadata about the resource |
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created |
  | 0..1 | code | Language of the resource content Binding: CommonLanguages (preferred) Max Binding: AllLanguages: A human language. | |
  | 0..1 | Narrative | Text summary of the resource, for human interpretation | |
  | 0..* | Resource | Contained, inline Resources | |
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |
  | 0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: ObservationCategoryCodes (extensible): Codes to denote a guideline or policy statement.when a genetic test result is being shared as a secondary finding. | |
  | 0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |
  | 0..1 | RelatedArtifact | Related Artifact URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/CGRelatedArtifact | |
  | ?! | 0..* | Extension | Extensions that cannot be ignored |
  | Σ | 0..* | Identifier | Business Identifier for observation |
  | Σ | 0..* | Reference(CarePlan | DeviceRequest | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | Fulfills plan, proposal or order |
  | Σ | 0..* | Reference(MedicationAdministration | MedicationDispense | MedicationStatement | Procedure | Immunization | ImagingStudy) | Part of referenced event |
  | ?!Σ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. |
  | 1..* | (Slice Definition) | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |
   | 1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |
    | Σ | 1..* | Coding | Code defined by a terminology system Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     | 0..1 | string | Version of the system - if relevant | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
     | 0..1 | string | Representation defined by the system | |
     | 0..1 | boolean | If this coding was chosen directly by the user | |
    | Σ | 0..1 | string | Plain text representation of the concept |
  | Σ | 1..1 | CodeableConcept | Type of observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. |
   | 0..1 | string | Unique id for inter-element referencing | |
   | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |
   | Σ | 0..* | Coding | Code defined by a terminology system |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |
    | Σ | 0..1 | uri | Identity of the terminology system Required Pattern: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes |
    | Σ | 0..1 | string | Version of the system - if relevant |
    | Σ | 0..1 | code | Symbol in syntax defined by the system Required Pattern: therapeutic-implication |
    | Σ | 0..1 | string | Representation defined by the system |
    | Σ | 0..1 | boolean | If this coding was chosen directly by the user |
   | Σ | 0..1 | string | Plain text representation of the concept |
  | Σ | 0..1 | Reference(Patient | Group | Location) | Who and/or what the observation is about |
  | Σ | 0..* | Reference(Resource) | What the observation is about, when it is not about the subject of record |
  | Σ | 0..1 | Reference(Encounter) | Healthcare event during which this observation is made |
  | Σ | 0..1 | Clinically relevant time/time-period for observation | |
   | dateTime | |||
   | Period | |||
   | Timing | |||
   | instant | |||
  | Σ | 0..1 | instant | Date/Time this version was made available |
  | Σ | 0..1 | Reference(Practitioner | PractitionerRole | Organization | CareTeam | Patient | RelatedPerson) | Who is responsible for the observation |
  | I | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
  | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
  | 0..* | Annotation | Comments about the observation | |
  | 0..1 | CodeableConcept | Observed body part Binding: SNOMEDCTBodyStructures (example): Codes describing anatomical locations. May include laterality. | |
  | 0..1 | CodeableConcept | How it was done Binding: ObservationMethods (example): Methods for simple observations. | |
  | 0..1 | Reference(Specimen) | Specimen used for this observation | |
  | 0..1 | Reference(Device | DeviceMetric) | (Measurement) Device | |
  | I | 0..* | BackboneElement | Provides guide for interpretation obs-3: Must have at least a low or a high or text |
   | 0..1 | string | Unique id for inter-element referencing | |
   | 0..* | Extension | Additional content defined by implementations | |
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
   | I | 0..1 | SimpleQuantity | Low Range, if relevant |
   | I | 0..1 | SimpleQuantity | High Range, if relevant |
   | 0..1 | CodeableConcept | Reference range qualifier Binding: ObservationReferenceRangeMeaningCodes (preferred): Code for the meaning of a reference range. | |
   | 0..* | CodeableConcept | Reference range population Binding: ObservationReferenceRangeAppliesToCodes (example): Codes identifying the population the reference range applies to. | |
   | 0..1 | Range | Applicable age range, if relevant | |
   | 0..1 | string | Text based reference range in an observation | |
  | Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Related resource that belongs to the Observation group |
  | Σ | 1..* | (Slice Definition) | Related measurements the observation is made from Slice: Unordered, Open by profile:$this.resolve() |
   | Σ | 0..* | Reference(Variant) | Related measurements the observation is made from |
   | Σ | 0..* | Reference(Genotype) | Related measurements the observation is made from |
   | Σ | 0..* | Reference(Haplotype) | Related measurements the observation is made from |
  | Σ | 0..* | (Slice Definition) | Component results Slice: Unordered, Open by pattern:code |
   | Content/Rules for all slices | |||
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. |
    | Σ | 0..1 | Actual component result | |
     | Quantity | |||
     | CodeableConcept | |||
     | string | |||
     | boolean | |||
     | integer | |||
     | Range | |||
     | Ratio | |||
     | SampledData | |||
     | time | |||
     | dateTime | |||
     | Period | |||
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Clinical conclusion (interpretation) of the observation |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 0..1 | string | Actual component result |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Level of Evidence |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL5356-2 (extensible) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Prognosis |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: prognostic-implication | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Associated phenotype |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81259-4 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Associated cancer |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-cancer | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Medication Assessed |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51963-7 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Associated cancer |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-therapy | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Effect medication metabolism |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53040-2 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL3856-3 (preferred) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Effect medication high risk |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 83009-1 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL2353-2 (extensible) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Effect medication high risk |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51961-1 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL539-8 (preferred) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
 Documentation for this format Documentation for this format | ||||
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | I | 0..* | GenomicImplication | Measurements and simple assertions |
 Documentation for this format Documentation for this format | ||||
This structure is derived from GenomicImplication
Differential View
This structure is derived from GenomicImplication
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | GenomicImplication | Measurements and simple assertions | |
  | ||||
   | ||||
    | 0..1 | uri | Identity of the terminology system Required Pattern: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
    | 0..1 | code | Symbol in syntax defined by the system Required Pattern: therapeutic-implication | |
  | 0..* | BackboneElement | Associated phenotype | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81259-4 | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..* | BackboneElement | Associated cancer | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-cancer | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..* | BackboneElement | Medication Assessed | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51963-7 | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..* | BackboneElement | Associated cancer | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-therapy | |
   | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined | |
  | 0..1 | BackboneElement | Effect medication metabolism | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53040-2 | |
   | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL3856-3 (preferred) | |
  | 0..1 | BackboneElement | Effect medication high risk | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 83009-1 | |
   | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL2353-2 (extensible) | |
  | 0..* | BackboneElement | Effect medication high risk | |
   | 1..1 | CodeableConcept | Type of component observation (code / type) Required Pattern: At least the following | |
    | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51961-1 | |
   | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL539-8 (preferred) | |
 Documentation for this format Documentation for this format | ||||
Snapshot View
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | I | 0..* | GenomicImplication | Measurements and simple assertions |
  | Σ | 0..1 | string | Logical id of this artifact |
  | Σ | 0..1 | Meta | Metadata about the resource |
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created |
  | 0..1 | code | Language of the resource content Binding: CommonLanguages (preferred) Max Binding: AllLanguages: A human language. | |
  | 0..1 | Narrative | Text summary of the resource, for human interpretation | |
  | 0..* | Resource | Contained, inline Resources | |
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |
  | 0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: ObservationCategoryCodes (extensible): Codes to denote a guideline or policy statement.when a genetic test result is being shared as a secondary finding. | |
  | 0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |
  | 0..1 | RelatedArtifact | Related Artifact URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/CGRelatedArtifact | |
  | ?! | 0..* | Extension | Extensions that cannot be ignored |
  | Σ | 0..* | Identifier | Business Identifier for observation |
  | Σ | 0..* | Reference(CarePlan | DeviceRequest | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | Fulfills plan, proposal or order |
  | Σ | 0..* | Reference(MedicationAdministration | MedicationDispense | MedicationStatement | Procedure | Immunization | ImagingStudy) | Part of referenced event |
  | ?!Σ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. |
  | 1..* | (Slice Definition) | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |
   | 1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |
    | Σ | 1..* | Coding | Code defined by a terminology system Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     | 0..1 | string | Version of the system - if relevant | |
     | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
     | 0..1 | string | Representation defined by the system | |
     | 0..1 | boolean | If this coding was chosen directly by the user | |
    | Σ | 0..1 | string | Plain text representation of the concept |
  | Σ | 1..1 | CodeableConcept | Type of observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. |
   | 0..1 | string | Unique id for inter-element referencing | |
   | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |
   | Σ | 0..* | Coding | Code defined by a terminology system |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |
    | Σ | 0..1 | uri | Identity of the terminology system Required Pattern: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes |
    | Σ | 0..1 | string | Version of the system - if relevant |
    | Σ | 0..1 | code | Symbol in syntax defined by the system Required Pattern: therapeutic-implication |
    | Σ | 0..1 | string | Representation defined by the system |
    | Σ | 0..1 | boolean | If this coding was chosen directly by the user |
   | Σ | 0..1 | string | Plain text representation of the concept |
  | Σ | 0..1 | Reference(Patient | Group | Location) | Who and/or what the observation is about |
  | Σ | 0..* | Reference(Resource) | What the observation is about, when it is not about the subject of record |
  | Σ | 0..1 | Reference(Encounter) | Healthcare event during which this observation is made |
  | Σ | 0..1 | Clinically relevant time/time-period for observation | |
   | dateTime | |||
   | Period | |||
   | Timing | |||
   | instant | |||
  | Σ | 0..1 | instant | Date/Time this version was made available |
  | Σ | 0..1 | Reference(Practitioner | PractitionerRole | Organization | CareTeam | Patient | RelatedPerson) | Who is responsible for the observation |
  | I | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
  | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
  | 0..* | Annotation | Comments about the observation | |
  | 0..1 | CodeableConcept | Observed body part Binding: SNOMEDCTBodyStructures (example): Codes describing anatomical locations. May include laterality. | |
  | 0..1 | CodeableConcept | How it was done Binding: ObservationMethods (example): Methods for simple observations. | |
  | 0..1 | Reference(Specimen) | Specimen used for this observation | |
  | 0..1 | Reference(Device | DeviceMetric) | (Measurement) Device | |
  | I | 0..* | BackboneElement | Provides guide for interpretation obs-3: Must have at least a low or a high or text |
   | 0..1 | string | Unique id for inter-element referencing | |
   | 0..* | Extension | Additional content defined by implementations | |
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
   | I | 0..1 | SimpleQuantity | Low Range, if relevant |
   | I | 0..1 | SimpleQuantity | High Range, if relevant |
   | 0..1 | CodeableConcept | Reference range qualifier Binding: ObservationReferenceRangeMeaningCodes (preferred): Code for the meaning of a reference range. | |
   | 0..* | CodeableConcept | Reference range population Binding: ObservationReferenceRangeAppliesToCodes (example): Codes identifying the population the reference range applies to. | |
   | 0..1 | Range | Applicable age range, if relevant | |
   | 0..1 | string | Text based reference range in an observation | |
  | Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Related resource that belongs to the Observation group |
  | Σ | 1..* | (Slice Definition) | Related measurements the observation is made from Slice: Unordered, Open by profile:$this.resolve() |
   | Σ | 0..* | Reference(Variant) | Related measurements the observation is made from |
   | Σ | 0..* | Reference(Genotype) | Related measurements the observation is made from |
   | Σ | 0..* | Reference(Haplotype) | Related measurements the observation is made from |
  | Σ | 0..* | (Slice Definition) | Component results Slice: Unordered, Open by pattern:code |
   | Content/Rules for all slices | |||
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. |
    | Σ | 0..1 | Actual component result | |
     | Quantity | |||
     | CodeableConcept | |||
     | string | |||
     | boolean | |||
     | integer | |||
     | Range | |||
     | Ratio | |||
     | SampledData | |||
     | time | |||
     | dateTime | |||
     | Period | |||
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Clinical conclusion (interpretation) of the observation |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 0..1 | string | Actual component result |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Level of Evidence |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL5356-2 (extensible) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Prognosis |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: prognostic-implication | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Associated phenotype |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 81259-4 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Associated cancer |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-cancer | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Medication Assessed |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51963-7 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Associated cancer |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/TbdCodes | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: associated-therapy | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: (unbound) (example): Binding not yet defined |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Effect medication metabolism |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 53040-2 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL3856-3 (preferred) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..1 | BackboneElement | Effect medication high risk |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 83009-1 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL2353-2 (extensible) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
   | Σ | 0..* | BackboneElement | Effect medication high risk |
    | 0..1 | string | Unique id for inter-element referencing | |
    | 0..* | Extension | Additional content defined by implementations | |
    | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized |
    | Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following |
     | 0..1 | string | Unique id for inter-element referencing | |
     | 0..* | Extension | Additional content defined by implementations | |
     | 1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      | 0..1 | string | Unique id for inter-element referencing | |
      | 0..* | Extension | Additional content defined by implementations | |
      | 1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      | 0..1 | string | Version of the system - if relevant | |
      | 1..1 | code | Symbol in syntax defined by the system Fixed Value: 51961-1 | |
      | 0..1 | string | Representation defined by the system | |
      | 0..1 | boolean | If this coding was chosen directly by the user | |
     | 0..1 | string | Plain text representation of the concept | |
    | Σ | 1..1 | CodeableConcept | Actual component result Binding: LOINC Answer List LL539-8 (preferred) |
    | I | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. |
    | 0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |
    | 0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |
 Documentation for this format Documentation for this format | ||||
Other representations of profile: Schematron
| Path | Conformance | ValueSet / Code |
| Observation.language | preferred | CommonLanguages Max Binding: AllLanguages |
| Observation.status | required | ObservationStatus |
| Observation.category | preferred | ObservationCategoryCodes |
| Observation.category:labCategory | preferred | ObservationCategoryCodes |
| Observation.code | example | LOINCCodes |
| Observation.dataAbsentReason | extensible | DataAbsentReason |
| Observation.interpretation | extensible | ObservationInterpretationCodes |
| Observation.bodySite | example | SNOMEDCTBodyStructures |
| Observation.method | example | ObservationMethods |
| Observation.referenceRange.type | preferred | ObservationReferenceRangeMeaningCodes |
| Observation.referenceRange.appliesTo | example | ObservationReferenceRangeAppliesToCodes |
| Observation.component.code | example | LOINCCodes |
| Observation.component.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:conclusion-string.code | example | Pattern: conclusion-string |
| Observation.component:conclusion-string.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:conclusion-string.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:evidence-level.code | example | Pattern: LOINC code 93044-6 |
| Observation.component:evidence-level.value[x] | extensible | LOINC LL5356-2 |
| Observation.component:evidence-level.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:evidence-level.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:prognosis.code | example | Pattern: prognostic-implication |
| Observation.component:prognosis.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:prognosis.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:associated-phenotype.code | example | Pattern: LOINC code 81259-4 |
| Observation.component:associated-phenotype.value[x] | example | |
| Observation.component:associated-phenotype.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:associated-phenotype.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:associated-cancer.code | example | Pattern: associated-cancer |
| Observation.component:associated-cancer.value[x] | example | |
| Observation.component:associated-cancer.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:associated-cancer.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:medication-assessed.code | example | Pattern: LOINC code 51963-7 |
| Observation.component:medication-assessed.value[x] | example | |
| Observation.component:medication-assessed.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:medication-assessed.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:therapy-assessed.code | example | Pattern: associated-therapy |
| Observation.component:therapy-assessed.value[x] | example | |
| Observation.component:therapy-assessed.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:therapy-assessed.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:effect-medication-metabolism.code | example | Pattern: LOINC code 53040-2 |
| Observation.component:effect-medication-metabolism.value[x] | preferred | LOINC LL3856-3 |
| Observation.component:effect-medication-metabolism.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:effect-medication-metabolism.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:effect-medication-high-risk.code | example | Pattern: LOINC code 83009-1 |
| Observation.component:effect-medication-high-risk.value[x] | extensible | LOINC LL2353-2 |
| Observation.component:effect-medication-high-risk.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:effect-medication-high-risk.interpretation | extensible | ObservationInterpretationCodes |
| Observation.component:effect-medication-efficacy.code | example | Pattern: LOINC code 51961-1 |
| Observation.component:effect-medication-efficacy.value[x] | preferred | LOINC LL539-8 |
| Observation.component:effect-medication-efficacy.dataAbsentReason | extensible | DataAbsentReason |
| Observation.component:effect-medication-efficacy.interpretation | extensible | ObservationInterpretationCodes |
| Id | Path | Details | Requirements |
| dom-2 | Observation | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | Observation | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | Observation | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | Observation | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | Observation | A resource should have narrative for robust management : text.`div`.exists() | |
| obs-6 | Observation | dataAbsentReason SHALL only be present if Observation.value[x] is not present : dataAbsentReason.empty() or value.empty() | |
| obs-7 | Observation | If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present : value.empty() or component.code.where(coding.intersect(%resource.code.coding).exists()).empty() | |
| ele-1 | Observation.meta | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.implicitRules | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.language | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.text | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.extension:secondaryfinding | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.extension:secondaryfinding | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.extension:bodyStructure | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.extension:bodyStructure | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.extension:relatedArtifact | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.extension:relatedArtifact | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.identifier | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.basedOn | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.partOf | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.status | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.category | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.category:labCategory | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.category:labCategory.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.category:labCategory.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.category:labCategory.coding | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.category:labCategory.text | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.code.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.code.coding | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code.coding.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.code.coding.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.code.coding.system | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code.coding.version | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code.coding.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code.coding.display | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code.coding.userSelected | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.code.text | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.subject | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.focus | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.encounter | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.effective[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.issued | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.performer | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.note | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.bodySite | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.method | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.specimen | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.device | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| obs-3 | Observation.referenceRange | Must have at least a low or a high or text : low.exists() or high.exists() or text.exists() | |
| ele-1 | Observation.referenceRange.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.referenceRange.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.referenceRange.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.referenceRange.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.referenceRange.low | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.referenceRange.high | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.referenceRange.type | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.referenceRange.appliesTo | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.referenceRange.age | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.referenceRange.text | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.hasMember | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.derivedFrom | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.derivedFrom:variant | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.derivedFrom:genotype | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.derivedFrom:haplotype | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:conclusion-string | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:conclusion-string.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:conclusion-string.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:conclusion-string.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:conclusion-string.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:conclusion-string.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:conclusion-string.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:conclusion-string.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:conclusion-string.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:conclusion-string.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:evidence-level | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:evidence-level.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:evidence-level.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:evidence-level.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:evidence-level.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:evidence-level.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:evidence-level.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:evidence-level.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:evidence-level.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:evidence-level.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:prognosis | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:prognosis.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:prognosis.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:prognosis.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:prognosis.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:prognosis.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:prognosis.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:prognosis.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:prognosis.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:prognosis.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-phenotype | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-phenotype.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:associated-phenotype.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:associated-phenotype.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:associated-phenotype.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:associated-phenotype.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-phenotype.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-phenotype.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-phenotype.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-phenotype.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-cancer | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-cancer.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:associated-cancer.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:associated-cancer.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:associated-cancer.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:associated-cancer.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-cancer.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-cancer.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-cancer.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:associated-cancer.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:medication-assessed | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:medication-assessed.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:medication-assessed.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:medication-assessed.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:medication-assessed.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:medication-assessed.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:medication-assessed.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:medication-assessed.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:medication-assessed.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:medication-assessed.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:therapy-assessed | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:therapy-assessed.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:therapy-assessed.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:therapy-assessed.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:therapy-assessed.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:therapy-assessed.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:therapy-assessed.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:therapy-assessed.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:therapy-assessed.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:therapy-assessed.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-metabolism | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-metabolism.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:effect-medication-metabolism.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:effect-medication-metabolism.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:effect-medication-metabolism.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:effect-medication-metabolism.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-metabolism.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-metabolism.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-metabolism.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-metabolism.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-high-risk | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-high-risk.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:effect-medication-high-risk.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:effect-medication-high-risk.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:effect-medication-high-risk.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:effect-medication-high-risk.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-high-risk.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-high-risk.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-high-risk.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-high-risk.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-efficacy | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-efficacy.extension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:effect-medication-efficacy.extension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:effect-medication-efficacy.modifierExtension | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | Observation.component:effect-medication-efficacy.modifierExtension | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| ele-1 | Observation.component:effect-medication-efficacy.code | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-efficacy.value[x] | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-efficacy.dataAbsentReason | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-efficacy.interpretation | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ele-1 | Observation.component:effect-medication-efficacy.referenceRange | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) |