This page is part of the Genetic Reporting Implementation Guide (v2.0.0: STU 2) based on FHIR (HL7® FHIR® Standard) R4. The current version which supersedes this version is 3.0.0. For a full list of available versions, see the Directory of published versions
This section provides a walk-through of mapping from an example Pharmacogenomics (PGx) report to FHIR using the structure definitions and guidance defined in this IG. The PGx report is represented as a Genomics Report. The related observations and other resources are available as links in the table at the bottom of this section. The JSON representation of this report can be found here.
A PGx report focuses on the impacts of a patient’s genetics to the behavior of medications, and can provide recommended adjustments to medication based on these impacts. The intention of this page is to provide a “User Guide”, based on a specific report example, highlighting how the defined profiles can be used together to encode important relations in PGx reporting. The report and images for this guide are built upon work done by the eMERGE (Electronic Medical Records and Genomics) program, funded by the National Human Genome Research Institute (NHGRI). The Clinical Genomics Working Group has worked closely with eMERGE to integrate community feedback from their initial mapping into this PGx user guide.
In order to receive a PGx report, you must first order a PGx test. Genomic tests, like all laboratory tests, are ordered using the Service Request resource. The example report in this section is based on a pre-coordinated gene panel defined by the sequencing laboratory. As such, the “code” element on the PGx report aligns with a pre-coordinated “code” element on the corresponding ServiceRequest, though the laboratory may provide additional codes as well. Additional structure may be in scope for the report if additional “orderDetail” data are present in the Service Request or if supporting information such as current medications are available during generation. An overview of the general relationships involved in ordering a report and receiving the results can be found here.
<ServiceRequest xmlns="http://hl7.org/fhir">
<id value="eMERGEServiceRequest"/>
...
<intent value="order"/>
<code>
<coding>
<system value="https://hgsc.bcm.edu/lab-test-codes/"/>
<code value="emerge-seq-ngs-pnl"/>
</coding>
</code>
<subject>
<reference value="Patient/CGPatientExample01"/>
</subject>
<specimen>
<reference value="Specimen/GenomicSpecimenExample01"/>
</specimen>
</ServiceRequest>
The guide is based on three figures. Figure 1 is an overview of the components of a PGx report. Figure 2 is a schema describing the use of the components from the HL7 Clinical Genomics WG Implementation Guide, and figure 3 is an example report with each element of the report mapped to the schema in figure 2. Use the numbers in each figure to find where the section/element goes. For example, #7 in figure 1 is information about the performing lab. In figure 2, #7 indicates an instance of the Organization Resource for the performer which would be a link through the performer element in the Genetics Diagnostic report. In figure 3, #7 indicates specific information about the performing lab found in the sample report. Thus, the specific performing lab information in figure 3 maps to an instance profile of Organization Resource linked to the Genetic Diagnostic report (#7 in figure 2).
In addition to the numbers, the colors in Figure 1 are intended to help guide the grouping of information in the report.
These images should provide you with a clear understanding of how the various FHIR PGx resources may be used when constructing a PGx report.
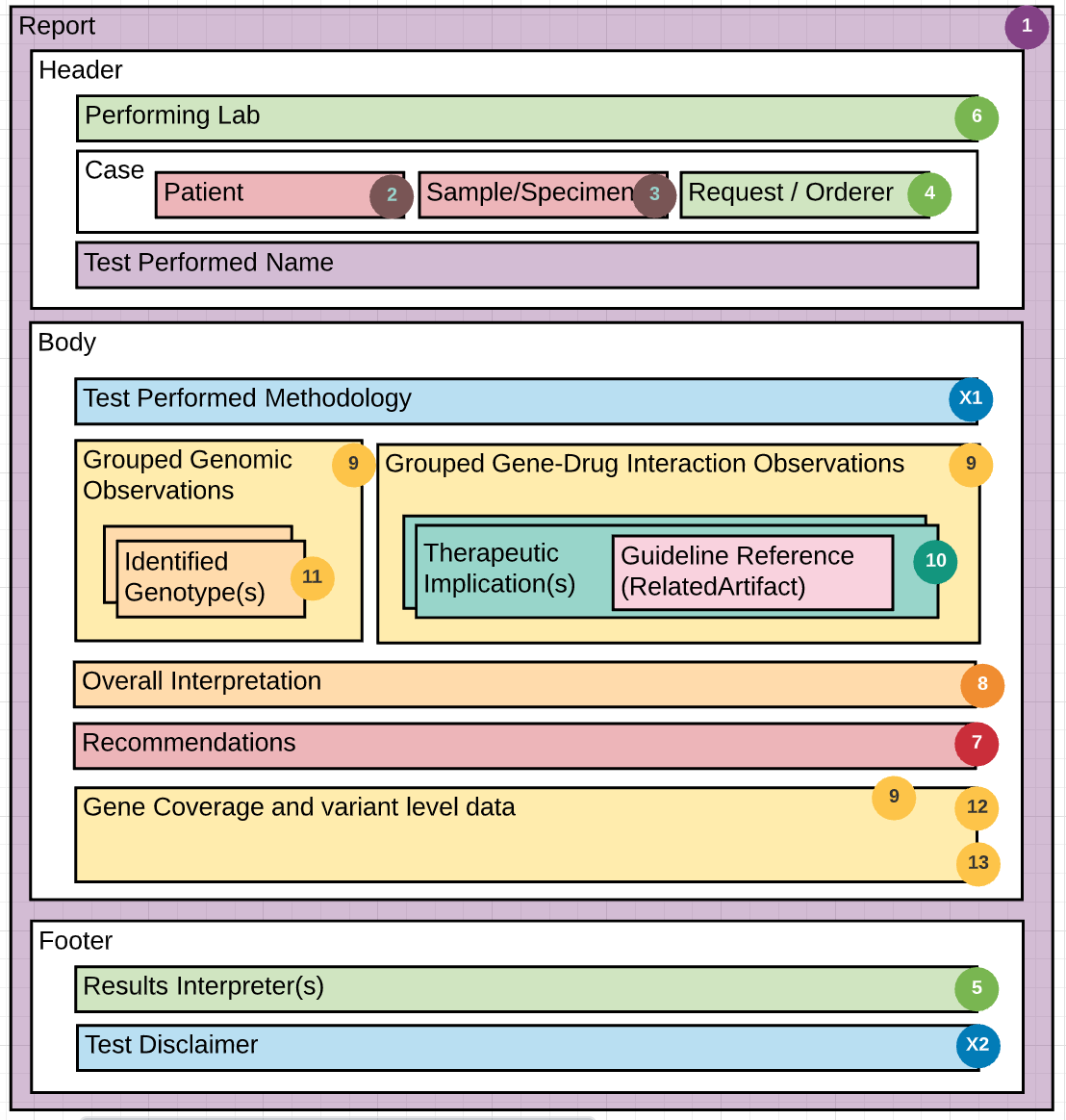
The Schema below depicts a “decomposition” of the PGx FHIR resources used to construct the example report. The color coding and numbers in the schema are mapped via the legend (image above) to the detailed PGx report shown in Figure #3.
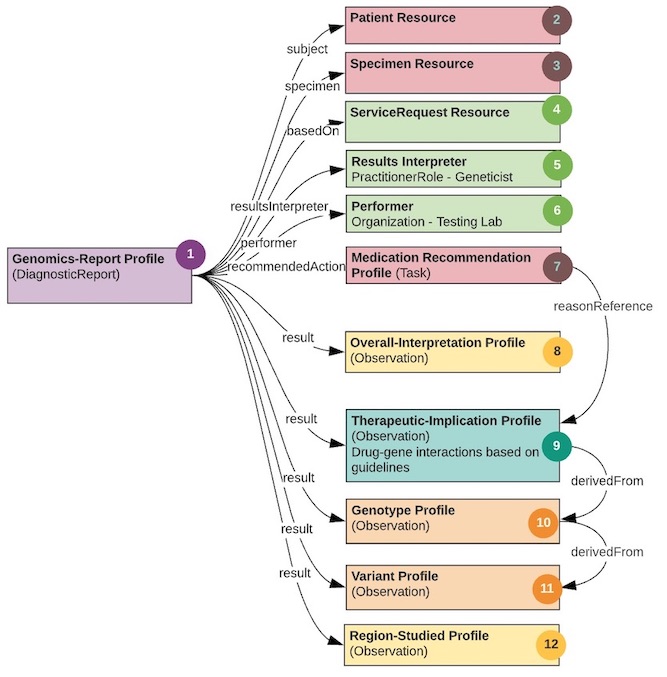
This PGx report, like all genomic reporting, is packaged with the diagnostic report profile. It conveys metadata about the overall report and carries multiple “result” Observations. All of the observations may “hang” directly off of the Genomics Report.
All pharmacogenomic implications are intended to be communicated using components within the Therapeutic Implication profile. The Medication-assessed component of the Therapeutic Implication profile, is the place to communicate the medication whose implication is being described, including a place for the coded value of the medication. Because this is an international profile, no guidance is provided on the medication coding system to be used. Instead, the appropriate code system for the jurisdiction, can and should be used.
To associate a Therapeutic-implication instance with a proposed recommended action (e.g., discontinuing a medication, altering dosage, or “choose alternative medication”) one can use the Medication Recommendation, potentially with a MedicationStatement instance. Use genomics-artifact in Therapeutic-implication to supply additional or supporting information, such as CPIC guidelines. In this report example, the text which explains the methodology of the test, was modeled using the PlanDefinition resource (#5 in the schema). The clinical genomics work group has not yet determined the value of providing a profile on PlanDefinition, and requests community feedback on encoding testing methodology details.
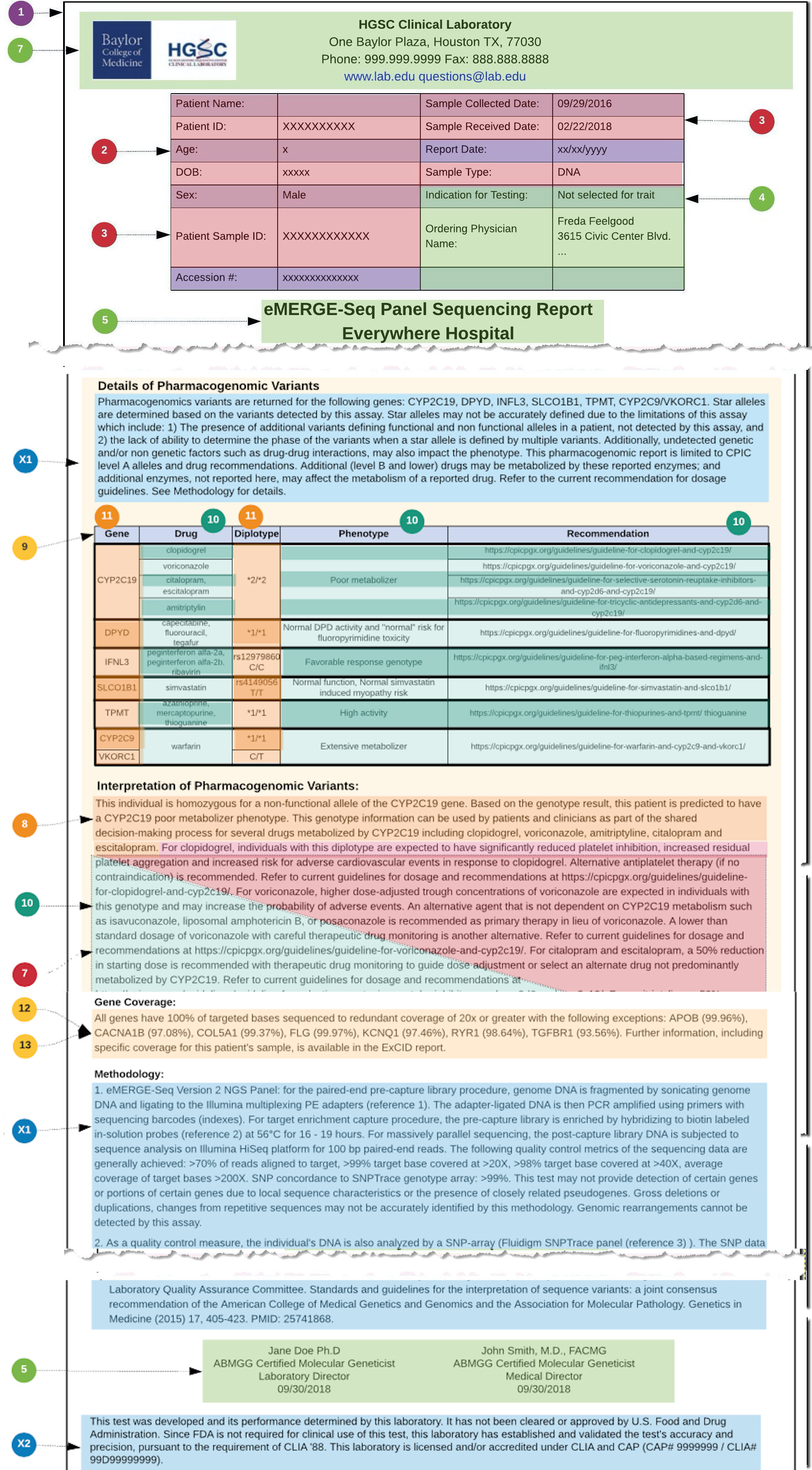
In the example used in this User Guide, the PGx results are based on diplotypes (star alleles), and the underlying variant observations, found in each relevant PGx gene that is covered by the PGx gene panel. These diplotypes are then used as a basis for relating PGx gene-drug knowledge implications. Note that there was also the choice in this use case, to derive Implication statements from one or more genotype Observations and not from variants directly.
Below is one of the therapeutic implications in the report:
<Observation xmlns="http://hl7.org/fhir">
...
<extension url="http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/CGRelatedArtifact">
<valueRelatedArtifact>
<type value="citation"/>
<url value="https://cpicpgx.org/guidelines/guideline-for-warfarin-and-cyp2c9-and-vkorc1/"/>
</valueRelatedArtifact>
</extension>
<code>
<coding>
<system value="http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs"/>
<code value="therapeutic-implication"/>
</coding>
</code>
<derivedFrom>
<reference value="Observation/Pgx-geno-1002"/>
<display value="CYP2C9*1/*1"/>
</derivedFrom>
<derivedFrom>
<reference value="Observation/Pgx-geno-1003"/>
<display value="VKORC1 rs9923231 C/T"/>
</derivedFrom>
<component>
<code>
<coding>
<system value="http://loinc.org"/>
<code value="51963-7"/>
</coding>
</code>
<valueCodeableConcept>
<coding>
<system value="http://ncimeta.nci.nih.gov"/>
<code value="C0043031"/>
<display value="warfarin"/>
</coding>
</valueCodeableConcept>
</component>
<component>
<code>
<coding>
<system value="http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs"/>
<code value="predicted-therapeutic-implication"/>
</coding>
</code>
<valueCodeableConcept>
<system value="http://loinc.org"/>
<code value="LA25391-6"/>
<display value="Normal Metabolizer"/>
</valueCodeableConcept>
</component>
<component>
<code>
<coding>
<system value="http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs"/>
<code value="conclusion-string"/>
</coding>
</code>
<valueString value="This individual is homozygous for the normal allele for the CYP2C9 gene. Based on the genotype result, this patient is predicted to have normal CYP2C9 function. This individual is also heterozygous for the variant allele for the VKORC1 gene. Expression level of the VKORC1 gene is associated with warfarin sensitivity. Based on the genotype result, this patient is predicted to have medium sensitivity to warfarin."/>
</component>
</Observation>
The following table contains additional information regarding the profiles used in mapping this example PGx report to FHIR, as well as individual example instances
| Profile | Description of PGx usage | Example links | Example Notes |
|---|---|---|---|
| Genomic Report | Intended to capture a single report, with a Code element aligning with a Service Request and multiple genomic result Observations. Not suitable for displaying summary information covering multiple reports. Extensions on DiagnosticReport can point to other reports. |
#DiagnosticReport-PGxGenomicsReportEMERGE.html | This report has results directly referenced and one Overall Interpretation |
| Overall genomic Interpretation | High-level observations of the result of the genomic testing, at the diagnostic report level. This is a flag often found as a statement in a report. For example, a report might begin ‘Positive - pathogenic variant found’ if a single pathogenic variant was found. The ‘positive’ trigger word in this statement is what would be captured. Again this is similar to a flag for an abnormal result found in laboratory messages. There is a new Observation component ‘conclusion-string’ which has been proposed to mirror the conclusion-string attribute on the DiagnosticReport, if additional detail is needed. One also has the option to use DiagnosticReport.text to provide overall information, but the ‘.text’ component would not call-out the conclusion. | #Observation-OverallInterpExample2.html | This example summarizes the potentially actionable results of the report. Codes should align with what is expected from the report’s ServiceRequest. |
| Genotype | Connects variant observations for homologous positions in the genome, or sets of homologous positions. In practice, it takes the capability of performing bioinformatic calculations to normalize across variant data to bring genotypes together. Genotype allows the sender to communicate a relationship between variant observations without the recipient needing to calculate the relationship. | #Observation-Pgx-geno-1001.html #Observation-Pgx-geno-1002.html #Observation-Pgx-geno-1003.html |
Genotype names such as star-alleles can appear in PGx reports either with or without a full description of the underlying observed variants. PGx guidance is often available at the level of these genotypes. |
| Variant | Intended for communication of positional information derived from the patient’s sample. The data should be suitable for calculations involving variant level details, such as clinical decision support or presence of a specific variant in a patient. Variant also carries important observational information about the occurrence of the variant in the patient/sample, and some pre-coordinated annotations such as gene name. More complex annotations such as pathogenicity should use the DiagnosticImplication profile for additional structure. Not all variants observed in a patient’s sample are required to be included in a report. |
#Observation-Pgx-var-1011.html #Observation-Pgx-var-1012.html #Observation-Pgx-var-1013.html #Observation-Pgx-var-1014.html #Observation-Pgx-var-1015.html #Observation-Pgx-var-1016.html #Observation-Pgx-var-1017.html #Observation-Pgx-var-1018.html |
In this example, variants are represented primarily using HGVS, with an Observation value of ‘Present’. This provides a computable observation of change, position in a reference sequence. Other components could be used to “define” the variant in question, for example ClinVar codes or a VCF column-like representation as seen in other examples. |
| Region Studied | The Region Studied profile is used to assert and describe actual regions studied for the performed test(s). Intended coverage areas may differ from actual coverage areas (e.g., due to technical limitations during test performance). This Observation does not carry a value. | #Observation-RegionStudiedPGx1.html #Observation-RegionStudiedPGx2.html #Observation-RegionStudiedPGx3.html |
These examples populate the gene-studied component and describe the coverage. Another approach would be to provide positional information on particular reference sequences. |
| Therapeutic Implication | All pharmacogenomic implications are intended to be communicated using components within the Therapeutic Implication profile, which has properties for observations that convey the potential impact of genomic characteristics on a medication or non-medicinal therapy. The medication-assessed component can be repeated to represent combination therapies. Otherwise, the recommendation is to limit Observations to one drug each to maintain semantic relationships in downstream systems. | #Observation-TxImp01.html #Observation-TxImp02.html #Observation-TxImp03.html #Observation-TxImp04.html #Observation-TxImp05.html |
In addition to the coded components, the conclusion-string component can provide text that the lab would normally include in a report narrative. Note that one implication can be derived from multiple genotypes and/or variant Observations, and those Observations may derive multiple implications. |
| Medication Recommendation | The Medication Recommendation is a recommended action Task describing what sort of change (if any) should be made for a specific medication based on an identified finding. This is a computable specific recommendation for action on a specific medication, such as changing a dose. It is a structured means of reporting recommendations. Here tied to a Therapeutic Implication to provide guidance such as changes to a dose or alternative medications. | #Task-PGxRecEx01.html #Task-PGxRecEx02.html #Task-PGxRecEx03.html #Task-PGxRecEx04.html #Task-PGxRecEx05.html |
Not included in this example, ‘Task.focus’ can be used to refer to a MedicationStatement for the patient, if available. See an alternative example using this pattern at #Task-MedicationRecommendationExample1.html. |
| Recommended Action (extension) | The recommended action (extension) references a proposed action that is recommended based on the results of the diagnostic report. It is not linked to individual variants and medications. Another possible type of recommendation is for additional follow-up testing of the subject or family members. This is an extension and may not be understood by all systems so care must be made that receivers are able to consume Task resources not normally linked with DiagnosticReports. It is intended as a textual recommendation. The information could also appear in the ‘text’ element of the diagnostic report, which may be improved to support coded data in future releases of FHIR. | Note that extensions may not be included in generated narrative summaries. | |
| Genomics Artifact (extension) | Captures citations, evidence and other supporting documentation for the observation or report. Included on the DiagnosticReport and Implication Observation structure through an extension. This dataType allows more structure than what would easily be represented in a text narrative. Useful for referring to guidelines, infobutton, lab methodology or any other supporting documentation. |
Note that extensions may not be included in generated narrative summaries. | |
| Genomics Risk Assessment | While not used in the eMERGE example, the Genomics Risk Assessment is used in a diagnostic report or observation to refer to a RiskAssessment resource. The RiskAssessment resource captures predicted outcomes for a patient or population on the basis of source information. Examples include: A prognosis statement for a particular condition; Risk of health outcome (heart attack, particular type of cancer) on the basis of lifestyle factors and/or family history; List of potential health risks based on a patient’s genetic analysis. This resource can be used to represent the results of formal scoring/decision support tools that evaluate risk. It can also be used to capture a practitioner’s subjective assessment of the patient’s risk based on existing knowledge and previous experience. |
Not used in this example | |
| Plan Definition | A plan definition is a pre-defined group of actions to be taken in particular circumstances, often including conditional elements, options, and other decision points. The resource is flexible enough to be used to represent a variety of workflows, as well as clinical decision support and quality improvement assets, including order sets, protocols, and decision support rules. | In the eMERGE report, this is used to provide additional methodology information, as an alternative to using related artifact or Observation.method. This use is still experimental and has not been included in our example. It is labelled as ‘X1’ in the figures. |
This Artifact List provides a comprehensive listing of the FHIR artifacts defined as part of the Genomics Implementation Guide and may be helpful when developing the PGx report.
Notable changes from the eMERGE III FHIR Template docs include:
For further information about the eMERGE Program, see: Pre-Print Journal Article: “Genomic Considerations for FHIR; eMERGE Implementation Lessons” and eMERGE Network Website.