This page is part of the Genetic Reporting Implementation Guide (v3.0.0-ballot: STU 3 Ballot 1) based on FHIR (HL7® FHIR® Standard) R4. The current version which supersedes this version is 3.0.0. For a full list of available versions, see the Directory of published versions
| Official URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study | Version: 3.0.0-ballot | |||
| Active as of 2023-12-18 | Computable Name: GenomicStudy | |||
A genomic study is a set of analyses performed to analyze and generate genomic data.
The details of a performed genomic study are captured as genomic study analysis instances, referenced via the genomic study's genomic-study-analysis-ext extension.
Genomic Study and Genomic Study Analysis profiles are backported from the FHIR R5 GenomicStudy resource.
A genomic study instance aims at delineating relevant information of a performed genomic study. A genomic study might comprise one or more analyses, each serving a specific purpose. These analyses may vary in method (e.g., karyotyping, CNV, or SNV detection), performer, software, devices used, or regions studied.
A genomic study instance describes relevant information including the reasons, purpose, and performers of the study. It also provides technical endpoints to access these data. It serves as a logical aggregator for complex genomic analyses.
A genomic study instance might be based on one or more service requests or care plans. The reasons for performing a genomic study might be driven by specific Conditions or Observations. Although the genomic study subject is the focus of the study, the analysis-level focus could be used to specify other relevant subjects or specimens. For example, trio testing may involve three subjects: the proband, and both parents. The proband would be the subject listed directly under the genomic study, while each of the analyses could represent proband, mother, and father genomic analyses.
In clinical use, the study interpreters use all the results of these various analyses to provide diagnostic and therapeutic insights for the patient, where performers are listed according to their participation in each genomic analysis. Each analysis may be based on a set of defined protocols that may differ from the performed protocols.
Detailed information about the results of the analyses may be represented by Observations and gathered in a Genomic Report. Both resources may refer back to the originating genomic study to allow navigation.
Since the release of R5 included the GenomicStudy resource, some concepts are being added and others are being slightly reframed. The Metrics extension is used to assert high level quality metrics. The Regions extension is used to assert details about the scope of the analysis. There are options to express detail about genes and regions in the analysis. These include:
When expressing the each of those elements, there are two approaches to consider:
The following figure is based on the Procedure/lungMass example. Some things to note:
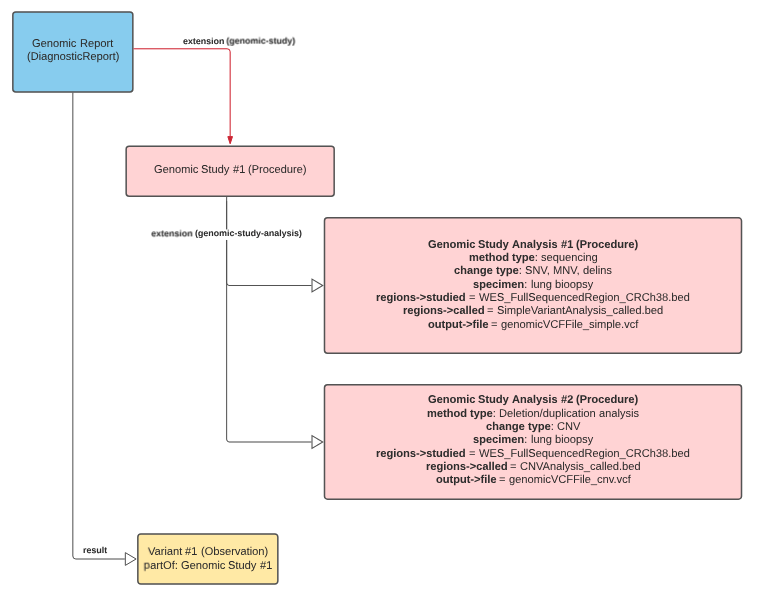
Usage:
Description of Profiles, Differentials, Snapshots and how the different presentations work.
This structure is derived from Procedure
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | Procedure | |||
  | 0..1 | Narrative | GenomicStudy.description | |
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |
  | 0..* | Reference(Genomic Study Analysis) | GenomicStudy.analysis URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-analysis-ext | |
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.referrer URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-referrer-ext | |
  | 0..* | Identifier | GenomicStudy.identifier | |
  | 0..* | canonical(PlanDefinition) | GenomicStudy.instantiatesCanonical | |
  | 0..* | uri | GenomicStudy.instantiatesUri | |
  | 0..* | Reference(ServiceRequest) | GenomicStudy.basedOn | |
  | 0..0 | |||
  | 1..1 | code | GenomicStudy.status | |
  | 0..0 | |||
  | 1..1 | CodeableConcept | Classification of the procedure | |
   | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
  | 0..1 | CodeableConcept | GenomicStudy.type Binding: Genomic Study Type ValueSet (example) | |
  | 1..1 | Reference(Patient | Group) | GenomicStudy.subject | |
  | 0..1 | Reference(Encounter) | GenomicStudy.encounter | |
  | 0..1 | dateTime | GenomicStudy.startDate | |
  | 0..0 | |||
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.interpreter | |
  | 0..0 | |||
  | 0..* | CodeableConcept | GenomicStudy.reason | |
  | 0..* | Reference(Condition | Observation) | GenomicStudy.reason | |
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..* | Annotation | GenomicStudy.note | |
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet |
| Procedure.code | example | GenomicStudyTypeVS (a valid code from Genomic Study Type CodeSystem) |
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Procedure | An action that is being or was performed on a patient | |
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created |
  | 0..1 | Narrative | GenomicStudy.description | |
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |
  | 0..* | Reference(Genomic Study Analysis) | GenomicStudy.analysis URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-analysis-ext | |
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.referrer URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-referrer-ext | |
  | ?! | 0..* | Extension | Extensions that cannot be ignored |
  | Σ | 0..* | Identifier | GenomicStudy.identifier |
  | Σ | 0..* | canonical(PlanDefinition) | GenomicStudy.instantiatesCanonical |
  | Σ | 0..* | uri | GenomicStudy.instantiatesUri |
  | Σ | 0..* | Reference(ServiceRequest) | GenomicStudy.basedOn |
  | ?!Σ | 1..1 | code | GenomicStudy.status Binding: EventStatus (required): A code specifying the state of the procedure. |
  | Σ | 1..1 | CodeableConcept | Classification of the procedure Binding: ProcedureCategoryCodes(SNOMEDCT) (example): A code that classifies a procedure for searching, sorting and display purposes. |
   | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
  | Σ | 0..1 | CodeableConcept | GenomicStudy.type Binding: Genomic Study Type ValueSet (example) |
  | Σ | 1..1 | Reference(Patient | Group) | GenomicStudy.subject |
  | Σ | 0..1 | Reference(Encounter) | GenomicStudy.encounter |
  | Σ | 0..1 | dateTime | GenomicStudy.startDate |
  | Σ | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.interpreter |
  | Σ | 0..* | CodeableConcept | GenomicStudy.reason Binding: ProcedureReasonCodes (example): A code that identifies the reason a procedure is required. |
  | Σ | 0..* | Reference(Condition | Observation) | GenomicStudy.reason |
  | 0..* | Annotation | GenomicStudy.note | |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet |
| Procedure.status | required | EventStatus |
| Procedure.category | example | ProcedureCategoryCodes(SNOMEDCT) |
| Procedure.code | example | GenomicStudyTypeVS (a valid code from Genomic Study Type CodeSystem) |
| Procedure.reasonCode | example | ProcedureReasonCodes |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Procedure | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Procedure | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Procedure | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() |
| Name | Flags | Card. | Type | Description & Constraints | ||||
|---|---|---|---|---|---|---|---|---|
 | 0..* | Procedure | An action that is being or was performed on a patient | |||||
  | Σ | 0..1 | id | Logical id of this artifact | ||||
  | Σ | 0..1 | Meta | Metadata about the resource | ||||
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  | 0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  | 0..1 | Narrative | GenomicStudy.description | |||||
  | 0..* | Resource | Contained, inline Resources | |||||
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
  | 0..* | Reference(Genomic Study Analysis) | GenomicStudy.analysis URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-analysis-ext | |||||
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.referrer URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-referrer-ext | |||||
  | ?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  | Σ | 0..* | Identifier | GenomicStudy.identifier | ||||
  | Σ | 0..* | canonical(PlanDefinition) | GenomicStudy.instantiatesCanonical | ||||
  | Σ | 0..* | uri | GenomicStudy.instantiatesUri | ||||
  | Σ | 0..* | Reference(ServiceRequest) | GenomicStudy.basedOn | ||||
  | ?!Σ | 1..1 | code | GenomicStudy.status Binding: EventStatus (required): A code specifying the state of the procedure. | ||||
  | Σ | 1..1 | CodeableConcept | Classification of the procedure Binding: ProcedureCategoryCodes(SNOMEDCT) (example): A code that classifies a procedure for searching, sorting and display purposes. | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
   | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
    | 0..1 | string | Version of the system - if relevant | |||||
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
    | 0..1 | string | Representation defined by the system | |||||
    | 0..1 | boolean | If this coding was chosen directly by the user | |||||
   | Σ | 0..1 | string | Plain text representation of the concept | ||||
  | Σ | 0..1 | CodeableConcept | GenomicStudy.type Binding: Genomic Study Type ValueSet (example) | ||||
  | Σ | 1..1 | Reference(Patient | Group) | GenomicStudy.subject | ||||
  | Σ | 0..1 | Reference(Encounter) | GenomicStudy.encounter | ||||
  | Σ | 0..1 | dateTime | GenomicStudy.startDate | ||||
  | Σ | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.interpreter | ||||
  | Σ | 0..* | BackboneElement | The people who performed the procedure | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations | |||||
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   | Σ | 0..1 | CodeableConcept | Type of performance Binding: ProcedurePerformerRoleCodes (example): A code that identifies the role of a performer of the procedure. | ||||
   | Σ | 1..1 | Reference(Practitioner | PractitionerRole | Organization | Patient | RelatedPerson | Device) | The reference to the practitioner | ||||
   | 0..1 | Reference(Organization) | Organization the device or practitioner was acting for | |||||
  | Σ | 0..* | CodeableConcept | GenomicStudy.reason Binding: ProcedureReasonCodes (example): A code that identifies the reason a procedure is required. | ||||
  | Σ | 0..* | Reference(Condition | Observation) | GenomicStudy.reason | ||||
  | 0..* | Annotation | GenomicStudy.note | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Conformance | ValueSet | ||||
| Procedure.language | preferred | CommonLanguages
| ||||
| Procedure.status | required | EventStatus | ||||
| Procedure.category | example | ProcedureCategoryCodes(SNOMEDCT) | ||||
| Procedure.code | example | GenomicStudyTypeVS (a valid code from Genomic Study Type CodeSystem) | ||||
| Procedure.performer.function | example | ProcedurePerformerRoleCodes | ||||
| Procedure.reasonCode | example | ProcedureReasonCodes | ||||
| Procedure.focalDevice.action | preferred | ProcedureDeviceActionCodes |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Procedure | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Procedure | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Procedure | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() |
This structure is derived from Procedure
Differential View
This structure is derived from Procedure
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | Procedure | |||
  | 0..1 | Narrative | GenomicStudy.description | |
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |
  | 0..* | Reference(Genomic Study Analysis) | GenomicStudy.analysis URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-analysis-ext | |
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.referrer URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-referrer-ext | |
  | 0..* | Identifier | GenomicStudy.identifier | |
  | 0..* | canonical(PlanDefinition) | GenomicStudy.instantiatesCanonical | |
  | 0..* | uri | GenomicStudy.instantiatesUri | |
  | 0..* | Reference(ServiceRequest) | GenomicStudy.basedOn | |
  | 0..0 | |||
  | 1..1 | code | GenomicStudy.status | |
  | 0..0 | |||
  | 1..1 | CodeableConcept | Classification of the procedure | |
   | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
  | 0..1 | CodeableConcept | GenomicStudy.type Binding: Genomic Study Type ValueSet (example) | |
  | 1..1 | Reference(Patient | Group) | GenomicStudy.subject | |
  | 0..1 | Reference(Encounter) | GenomicStudy.encounter | |
  | 0..1 | dateTime | GenomicStudy.startDate | |
  | 0..0 | |||
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.interpreter | |
  | 0..0 | |||
  | 0..* | CodeableConcept | GenomicStudy.reason | |
  | 0..* | Reference(Condition | Observation) | GenomicStudy.reason | |
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
  | 0..* | Annotation | GenomicStudy.note | |
  | 0..0 | |||
  | 0..0 | |||
  | 0..0 | |||
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet |
| Procedure.code | example | GenomicStudyTypeVS (a valid code from Genomic Study Type CodeSystem) |
Key Elements View
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | 0..* | Procedure | An action that is being or was performed on a patient | |
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created |
  | 0..1 | Narrative | GenomicStudy.description | |
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |
  | 0..* | Reference(Genomic Study Analysis) | GenomicStudy.analysis URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-analysis-ext | |
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.referrer URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-referrer-ext | |
  | ?! | 0..* | Extension | Extensions that cannot be ignored |
  | Σ | 0..* | Identifier | GenomicStudy.identifier |
  | Σ | 0..* | canonical(PlanDefinition) | GenomicStudy.instantiatesCanonical |
  | Σ | 0..* | uri | GenomicStudy.instantiatesUri |
  | Σ | 0..* | Reference(ServiceRequest) | GenomicStudy.basedOn |
  | ?!Σ | 1..1 | code | GenomicStudy.status Binding: EventStatus (required): A code specifying the state of the procedure. |
  | Σ | 1..1 | CodeableConcept | Classification of the procedure Binding: ProcedureCategoryCodes(SNOMEDCT) (example): A code that classifies a procedure for searching, sorting and display purposes. |
   | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following |
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
  | Σ | 0..1 | CodeableConcept | GenomicStudy.type Binding: Genomic Study Type ValueSet (example) |
  | Σ | 1..1 | Reference(Patient | Group) | GenomicStudy.subject |
  | Σ | 0..1 | Reference(Encounter) | GenomicStudy.encounter |
  | Σ | 0..1 | dateTime | GenomicStudy.startDate |
  | Σ | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.interpreter |
  | Σ | 0..* | CodeableConcept | GenomicStudy.reason Binding: ProcedureReasonCodes (example): A code that identifies the reason a procedure is required. |
  | Σ | 0..* | Reference(Condition | Observation) | GenomicStudy.reason |
  | 0..* | Annotation | GenomicStudy.note | |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet |
| Procedure.status | required | EventStatus |
| Procedure.category | example | ProcedureCategoryCodes(SNOMEDCT) |
| Procedure.code | example | GenomicStudyTypeVS (a valid code from Genomic Study Type CodeSystem) |
| Procedure.reasonCode | example | ProcedureReasonCodes |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Procedure | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Procedure | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Procedure | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() |
Snapshot View
| Name | Flags | Card. | Type | Description & Constraints | ||||
|---|---|---|---|---|---|---|---|---|
 | 0..* | Procedure | An action that is being or was performed on a patient | |||||
  | Σ | 0..1 | id | Logical id of this artifact | ||||
  | Σ | 0..1 | Meta | Metadata about the resource | ||||
  | ?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  | 0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  | 0..1 | Narrative | GenomicStudy.description | |||||
  | 0..* | Resource | Contained, inline Resources | |||||
  | 0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
  | 0..* | Reference(Genomic Study Analysis) | GenomicStudy.analysis URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-analysis-ext | |||||
  | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.referrer URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/genomic-study-referrer-ext | |||||
  | ?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  | Σ | 0..* | Identifier | GenomicStudy.identifier | ||||
  | Σ | 0..* | canonical(PlanDefinition) | GenomicStudy.instantiatesCanonical | ||||
  | Σ | 0..* | uri | GenomicStudy.instantiatesUri | ||||
  | Σ | 0..* | Reference(ServiceRequest) | GenomicStudy.basedOn | ||||
  | ?!Σ | 1..1 | code | GenomicStudy.status Binding: EventStatus (required): A code specifying the state of the procedure. | ||||
  | Σ | 1..1 | CodeableConcept | Classification of the procedure Binding: ProcedureCategoryCodes(SNOMEDCT) (example): A code that classifies a procedure for searching, sorting and display purposes. | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
   | Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
    | 0..1 | string | Unique id for inter-element referencing | |||||
    | 0..* | Extension | Additional content defined by implementations | |||||
    | 1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
    | 0..1 | string | Version of the system - if relevant | |||||
    | 1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
    | 0..1 | string | Representation defined by the system | |||||
    | 0..1 | boolean | If this coding was chosen directly by the user | |||||
   | Σ | 0..1 | string | Plain text representation of the concept | ||||
  | Σ | 0..1 | CodeableConcept | GenomicStudy.type Binding: Genomic Study Type ValueSet (example) | ||||
  | Σ | 1..1 | Reference(Patient | Group) | GenomicStudy.subject | ||||
  | Σ | 0..1 | Reference(Encounter) | GenomicStudy.encounter | ||||
  | Σ | 0..1 | dateTime | GenomicStudy.startDate | ||||
  | Σ | 0..1 | Reference(Practitioner | PractitionerRole) | GenomicStudy.interpreter | ||||
  | Σ | 0..* | BackboneElement | The people who performed the procedure | ||||
   | 0..1 | string | Unique id for inter-element referencing | |||||
   | 0..* | Extension | Additional content defined by implementations | |||||
   | ?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   | Σ | 0..1 | CodeableConcept | Type of performance Binding: ProcedurePerformerRoleCodes (example): A code that identifies the role of a performer of the procedure. | ||||
   | Σ | 1..1 | Reference(Practitioner | PractitionerRole | Organization | Patient | RelatedPerson | Device) | The reference to the practitioner | ||||
   | 0..1 | Reference(Organization) | Organization the device or practitioner was acting for | |||||
  | Σ | 0..* | CodeableConcept | GenomicStudy.reason Binding: ProcedureReasonCodes (example): A code that identifies the reason a procedure is required. | ||||
  | Σ | 0..* | Reference(Condition | Observation) | GenomicStudy.reason | ||||
  | 0..* | Annotation | GenomicStudy.note | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Conformance | ValueSet | ||||
| Procedure.language | preferred | CommonLanguages
| ||||
| Procedure.status | required | EventStatus | ||||
| Procedure.category | example | ProcedureCategoryCodes(SNOMEDCT) | ||||
| Procedure.code | example | GenomicStudyTypeVS (a valid code from Genomic Study Type CodeSystem) | ||||
| Procedure.performer.function | example | ProcedurePerformerRoleCodes | ||||
| Procedure.reasonCode | example | ProcedureReasonCodes | ||||
| Procedure.focalDevice.action | preferred | ProcedureDeviceActionCodes |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Procedure | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Procedure | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Procedure | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Procedure | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() |
This structure is derived from Procedure
Other representations of profile: CSV, Excel, Schematron