|
Type
|
Reference
|
Content
|
|
web
|
github.com
|
For example, a medication history reported directly by a patient may not carry the same evidentiary weight as a medication order formally documented by a prescribing clinician. To address this limitation, the Implementation Guide recommends using OMOP’s observation_type_concept_id or drug_type_concept_id fields to indicate the provenance of each record. (See Type Concepts in the OMOP Common Data Model
) By explicitly tagging records with their source, implementers can support analyses that require higher confidence in data accuracy or that need to filter data based on verification status. This practice improves transparency and helps maintain analytic rigor in research contexts where the reliability of the underlying data is critical.
|
|
web
|
athena.ohdsi.org
|
A Standard Concept representing the disease phase, outcome, or other abstraction of which the episode consists. For example, if the EPISODE_CONCEPT_ID is treatment regimen
then the EPISODE_OBJECT_CONCEPT_ID should contain the chemotherapy regimen concept, like Afatinib monotherapy
.
|
|
web
|
athena.ohdsi.org
|
A Standard Concept representing the disease phase, outcome, or other abstraction of which the episode consists. For example, if the EPISODE_CONCEPT_ID is treatment regimen
then the EPISODE_OBJECT_CONCEPT_ID should contain the chemotherapy regimen concept, like Afatinib monotherapy
.
|
|
web
|
athena.ohdsi.org
|
The FACT_RELATIONSHIP table contains records about the relationships between facts stored as records in any table of the CDM. Relationships can be defined between facts from the same domain, or different domains. Examples of Fact Relationships include: Person relationships
(parent-child), care site relationships (hierarchical organizational structure of facilities within a health system), indication relationship (between drug exposures and associated conditions), usage relationships (of devices during the course of an associated procedure), or facts derived from one another (measurements derived from an associated specimen).
|
|
web
|
athena.ohdsi.org
|
The Concept Id representing the country. Values should conform to the Geography
domain.
|
|
web
|
athena.ohdsi.org
|
The meaning of Concept 4172703
for '=' is identical to omission of a OPERATOR_CONCEPT_ID value. Since the use of this field is rare, it's important when devising analyses to not to forget testing for the content of this field for values different from =.
|
|
web
|
ucum.org
|
FHIR Observation units are embedded within the Quantity type structure, typically utilizing standardized systems such as UCUM (Unified Code for Units of Measure)
. The mapping process must preserve unit information through OMOP's unit_concept_id
field in the Measurement table.
|
|
web
|
www.researchallofus.org
|
All collected data are maintained and curated by the All of Us Data and Research Center (DRC)
and are accessible to researchers through the All of Us Researcher Workbench
. The primary data standard and clinical data model utilized by the All of Us Research Program is OMOP.
|
|
web
|
doi.org
|
Renedo, D., Acosta, J. N., Sujijantarat, N., Antonios, J. P., et al. (2022)
. Carotid Artery Disease Among Broadly Defined Underrepresented Groups: The All of Us Research Program.
Stroke, 53. https://doi.org/10.1161/STROKEAHA.121.037554
|
|
web
|
www.ahajournals.org
|
Data Supplement
|
|
web
|
doi.org
|
Khan, M. S., Carroll, R. J. (2022)
. Inference-based correction of multi-site height and weight measurement data in the All of Us Research Program.
Journal of the American Medical Informatics Association, 29(4), 626–630. https://doi.org/10.1093/jamia/ocab251
|
|
web
|
academic.oup.com
|
Data Supplement
(additional OMOP concept IDs included in article text)
|
|
web
|
doi.org
|
Berman, L., Ostchega, Y., Giannini, J., Anandan, L. P., et al. (2024)
. Application of a Data Quality Framework to Ductal Carcinoma In Situ Using Electronic Health Record Data From the All of Us Research Program.
JCO Clinical Cancer Informatics. https://doi.org/10.1200/CCI.24.00052
|
|
web
|
ascopubs.org
|
Data Supplement
|
|
web
|
ohdsi.github.io
|
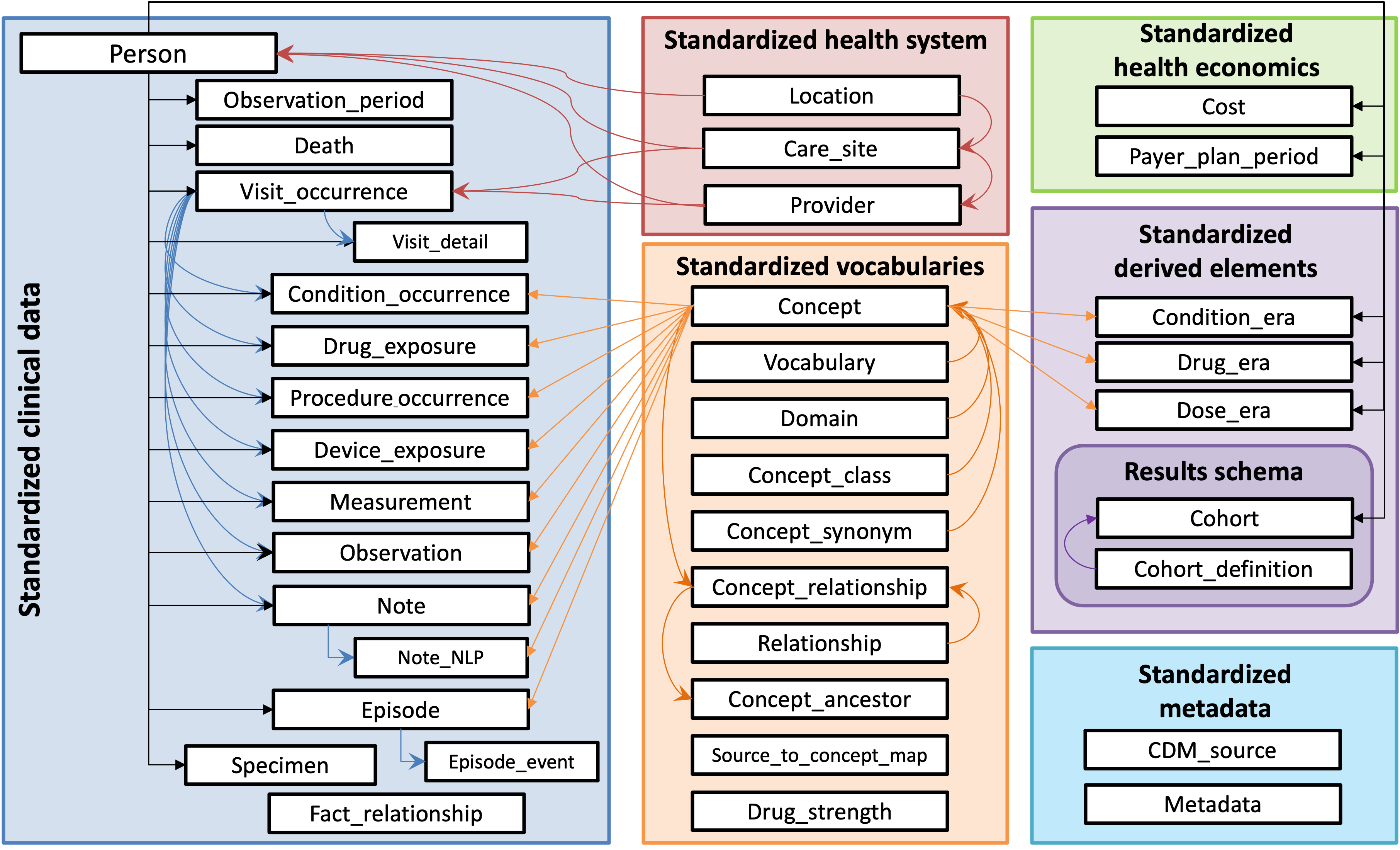
The OMOP CDM is designed for large‑scale aggregation, whether in centralized repositories or distributed research networks. Its domain structure and Standardized Vocabularies allow data from heterogeneous sources—each with distinct schemas and code systems—to be normalized into a single analytic framework. Every concept is assigned a status of Standard
, Non‑Standard
, or Classification
; only one Standard concept exists for each clinical idea within a domain, eliminating ambiguity and enabling cross‑site comparability. (See: OMOP Vocabulary Concept Structure
for additional details)
|
|
web
|
ohdsi.github.io
|
Because a single FHIR resource may contains multiple clinical ideas, each coded element must be evaluated independently. A FHIR Observation may yield Measurement, Condition Occurrence, or Procedure Occurrence records in OMOP, depending on the specific codes it carries. Terminologies commonly used in FHIR—such as LOINC, SNOMED CT, and RxNorm—frequently align with OMOP Standard concepts. In cases where no Standard mapping exists, particularly in cases such as HL7‑maintained value sets, or local codes, a extension concepts may be added to the OMOP concept table with concept_id
values of 2,000,000,000 (2 billion) or higher may be created. These “2-billionaires” IDs preserve information without loss and can be retired once the community adopts an official Standard concept. Regular vocabulary updates are therefore essential, both for pipelines ingesting new data and for legacy OMOP datastores. (See: OMOP CDM: Custom Concepts
for more information)
|
|
web
|
athena.ohdsi.org
|
The OHDSI Athena
website provides both access to request OHDSI Vocabulary downloads and a comprehensive searchable database that serves dual purposes for mapping activities. Implementers can use this resource to manually browse available vocabularies and identify codes that appropriately match source concepts to Standard OMOP concepts and also receive vocabulary updates when new versions of the OHDSI Stabdardized Vocabularies are published. Utilization of the OHDSI Standardized Vocabularies is essential for validating mappings and resolving complex terminology translation challenges that arise during FHIR to OMOP transformation.
|
|
web
|
github.com
|
Translate source code to concept ID: Utilise the ConceptMap/$translate
FHIR terminology server operation.
|
|
web
|
github.com
|
Lookup concept properties: Utilise the CodeSystem/$lookup
FHIR terminology server operation.
|
|
web
|
athena.ohdsi.org
|
Note the table below is only a partial list. A complete listing of type concepts can be found in the OHDSI Standardized Vocabularies
and a detailed explanation is available on the OHDSI Vocabulary wiki
.
|
|
web
|
github.com
|
Note the table below is only a partial list. A complete listing of type concepts can be found in the OHDSI Standardized Vocabularies
and a detailed explanation is available on the OHDSI Vocabulary wiki
.
|
|
web
|
ohdsi.github.io
|
Drug Exposure
|
|
web
|
ohdsi.github.io
|
Condition Occurrence
|
|
web
|
ohdsi.github.io
|
Procedure Occurrence
|
|
web
|
ohdsi.github.io
|
Visit Occurrence
|
|
web
|
ohdsi.github.io
|
Measurement
|
|
web
|
ohdsi.github.io
|
Observation
|
|
web
|
www.hl7vulcan.org
|
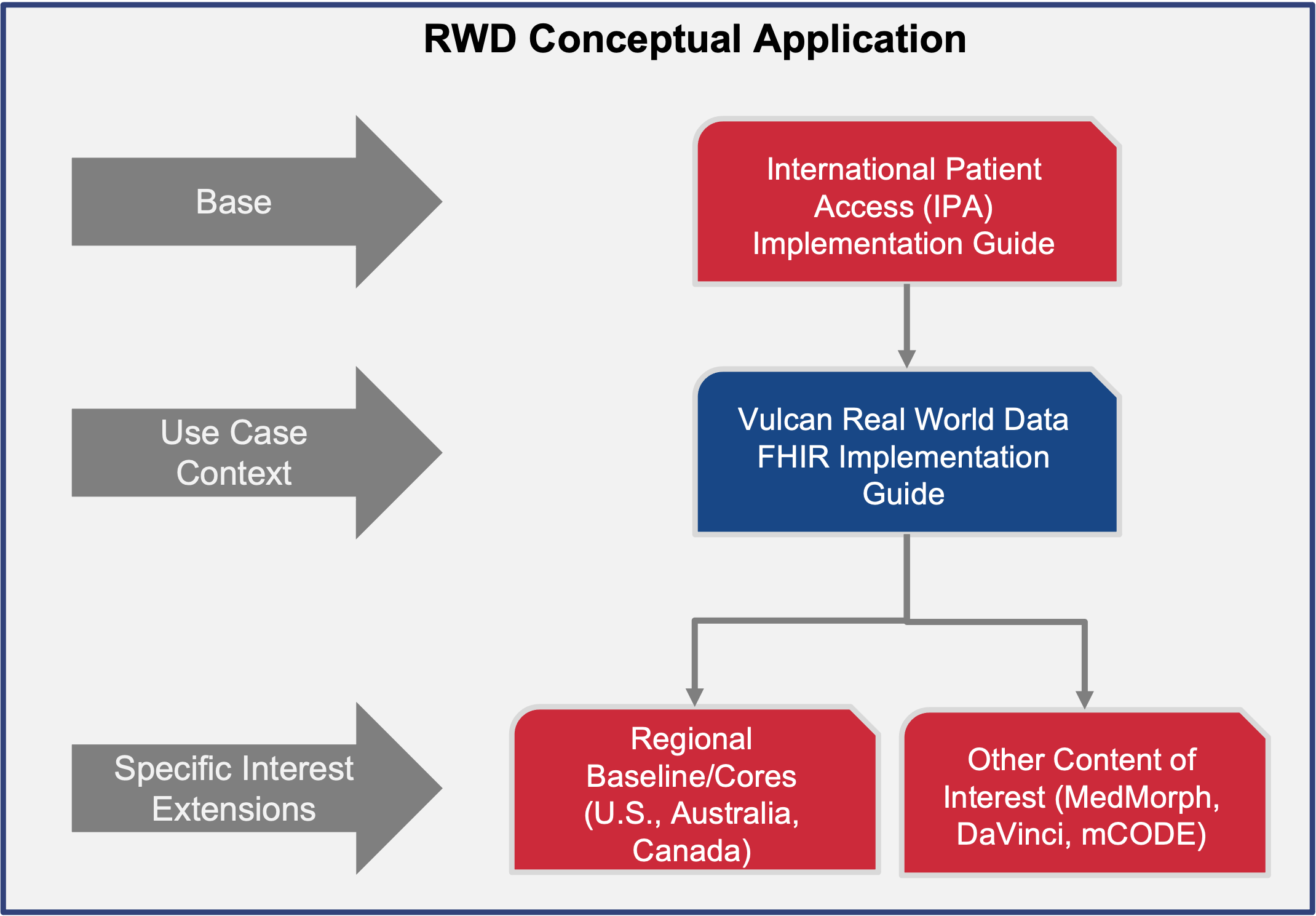
This Implementation Guide was developed under the auspices of the Vulcan
FHIR to OMOP work group in collaboration with the OHDSI OMOP + FHIR working group
.
|
|
web
|
teams.microsoft.com
|
This Implementation Guide was developed under the auspices of the Vulcan
FHIR to OMOP work group in collaboration with the OHDSI OMOP + FHIR working group
.
|
|
web
|
www.hl7vulcan.org
|
FHIR benefits from ongoing enhancements through the efforts of the HL7 Biomedical Research and Regulation (BR&R) Working Group
and the Vulcan FHIR Accelerator
supporting data exchange for real world data in observational and translational research. Whereas the FHIR accelerators and its user-community at-large are achieving enormous strides developing focused utilization of the standard in many key areas, FHIR developers do not aim to develop analytic methods that generate evidence at-scale as do members of the OHDSI community. Rather, FHIR’s role in a research data ecosystem is provision of a platform capable of supporting lossless exchange of data, knowledge and other health artifacts via its numerous FHIR resources and compliant APIs. In contrast, the OMOP Common Data Model was designed to store observational data optimized for research analytics and for utilization by the OHDSI community using OMOP-compliant tools to generate novel research methods and discoveries improving health. FHIR and OMOP serve complementary but distinct purposes making harmonization between them essential for effective data interoperability. Implementation guides for transforming between these standards address the core interoperability paradox where "more standards lead to less interoperability," by providing standardized pathways that minimize information loss during transformation processes.[6]
|
|
web
|
colab.research.google.com
|
Created to support the Vulcan July 2025 Connectathon, this Jupyter notebook
provides basic syntax and specification validation of FHIR JSON and OMOP CSV data for the Vulcan FHIR->OMOP Connectathon. The notebook can be run as-is from within Google Colab (with a Google account) or downloaded and run locally. Upload your FHIR JSON or OMOP CSVs into the colab temporary directory and update the paths accordingly.
|