This page is part of the Vulcan FHIR to OMOP FHIR Implementation Guide (v1.0.0-ballot: INFORMATIVE 1 Ballot 1) based on FHIR (HL7® FHIR® Standard) v5.0.0. No current official version has been published yet. For a full list of available versions, see the Directory of published versions
| Page standards status: Informative |
The canonical use case employed is querying a FHIR-enabled data repository to retrieve records and load them into an OMOP data store. This guide defines a set of logical models that represent the OMOP Common Data Model, v5.4. It also provides mappings, defined as FHIR StructureMaps and ConceptMaps (hosted on a FHIR terminology server), between the International Patient Access FHIR and US Core Encounter and Procedure profiles and the OMOP data tables.
The NIH's All of Us Research Program is a historic initiative aiming to collect and analyze data from one million or more individuals residing in the United States. Currently, the Program has enrolled over 849,000 participants. The primary goal of the initiative is to promote better health outcomes by empowering thousands of researchers with diverse, longitudinal data gathered from participants—80% of whom are traditionally underrepresented in biomedical research.
The Program collects various data types from consented participants, including:
Additional data currently being linked to participant records include:
All collected data are maintained and curated by the All of Us Data and Research Center (DRC) and are accessible to researchers through the All of Us Researcher Workbench. The primary data standard and clinical data model utilized by the All of Us Research Program is OMOP.
The All of Us Program collaborates with approximately 50 Health Provider Organizations (HPOs) across the United States to recruit participants. The All of Us Research Program leverages OMOP to standardize and structure its collected data, ensuring feasibility and facilitating researchers' ability to access and integrate data from diverse sources. These HPO partners extract participant EHR data, convert it into OMOP format, and submit 22 OMOP tables to the DRC on a quarterly basis. Given that EHR data represent the greatest demand for FHIR to OMOP mapping among the various All of Us data types, the All of Us program exemplifies real-world use cases for the utilization of EHR-sourced FHIR data transformed onto the OMOP CDM to generate evidence.
Studies were selected as examples based on the following criteria:
These selected studies offer practical, real-world examples of codes, data elements, and concept IDs. It is important to note that the examples may also incorporate additional codes and OMOP concept IDs as required.
The Vulcan Retrieval of Real World Data (RWD) for Clinical Research Implementation Guide (IG) aims to facilitate the transmission of clinical data generated during routine patient care (Electronic Health Record data, EHR) for use as Real World Data (RWD) supporting clinical research. Traditionally, clinical trials rely on prospective data collection tailored to specific research questions and analytics. In contrast, data generated through routine healthcare operations are rarely aligned with secondary research requirements. The Vulcan RWD IG defines FHIR profiles suitable for use within EHR systems, facilitating the retrieval and alignment of relevant clinical data from RWD sources for clinical research and regulatory submissions.
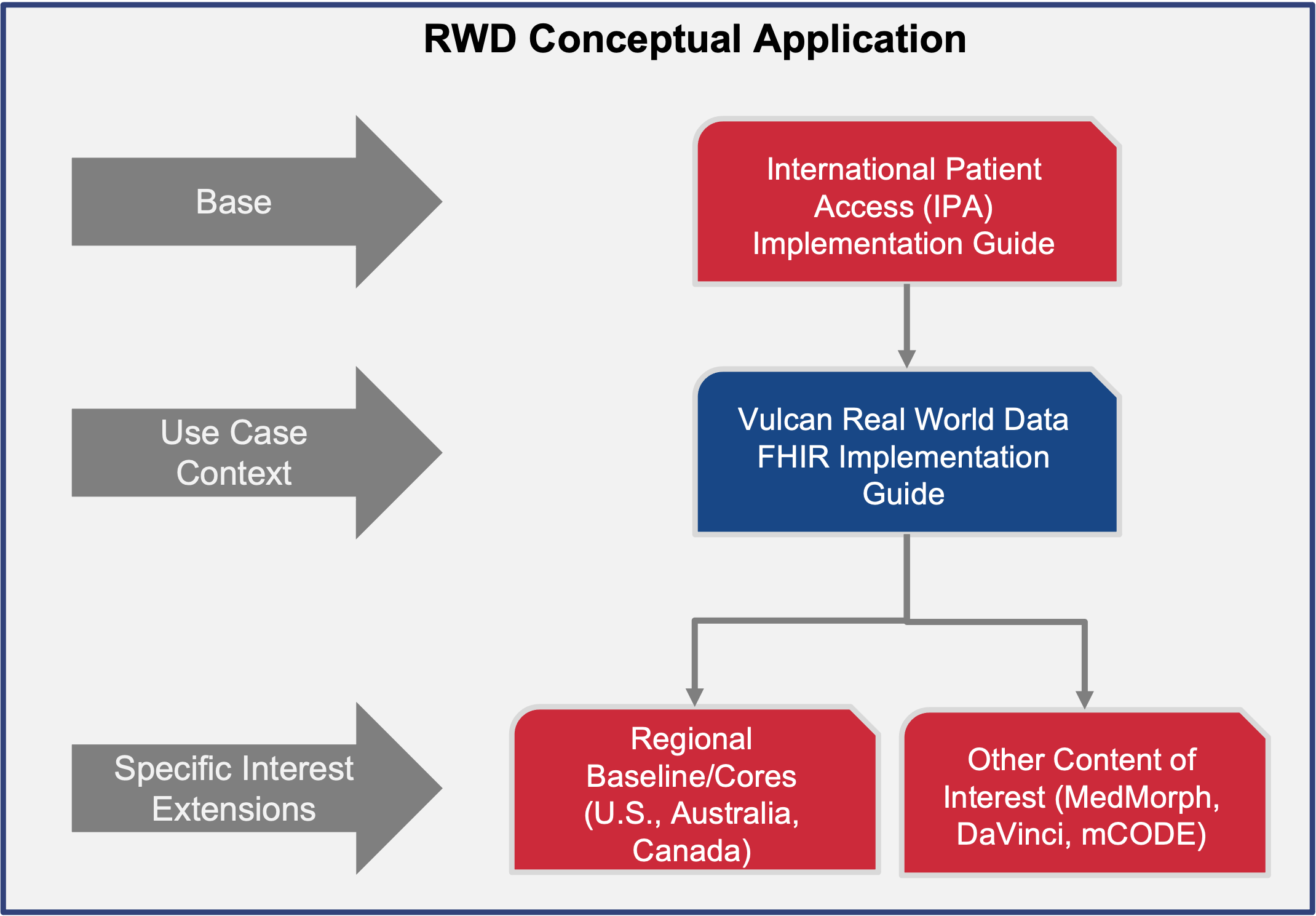
The Vulcan Real-World Data IG leverages the FHIR International Patient Access (IPA) IG to provide a foundational dataset and profiles necessary to address RWD use case requirements.
These studies referenced in the Vulcan RWD IG specify the data elements required to identify, request, and retrieve relevant EHR data aligned with each study’s cohort definition and research parameters. Most data elements are derived from the IPA profiles, providing a standardized minimal dataset. The Vulcan RWD IG further supports flexibility to accommodate local, regional, or study-specific needs through extensions, as illustrated in the RWD Conceptual Application Model pictured below:
The healthcare industry is increasingly leveraging Artificial Intelligence (AI) to extract actionable insights from clinical data. The OMOP Common Data Model is widely used for AI model training due to its robust, standardized structure designed to support large-scale data analysis, including clinical outcomes, drug safety, and efficacy research. However, FHIR (Fast Healthcare Interoperability Resources) is a newer, more flexible standard for health information exchange and is widely adopted for its ability to support real-time clinical workflows, such as through electronic health records (EHRs).
While OMOP provides a reliable foundation for building AI models, FHIR's versatility and growing adoption make it a popular choice for applying AI, which presents challenges when it comes to directly applying OMOP-trained AI models trained to FHIR data. The key issue is that AI models typically require data in a highly structured format (such as OMOP), but FHIR resources are more granular and often transmitted in a fragmented or single-message form. This misalignment makes it difficult to directly use FHIR for training AI models, which is why a transformation from FHIR to OMOP is necessary.
To bridge this gap, the use of Extract, Transform, Load (ETL) processes to convert FHIR data into OMOP format has emerged as a critical solution. By transforming FHIR resources—whether from a single message (such as a single patient encounter or medication prescription) or aggregated messages—into the standardized OMOP format, healthcare organizations can train AI models on OMOP data while leveraging real-time clinical data in FHIR for classification tasks.
The transformation enables AI models trained on the OMOP CDM to be applied to clinical data captured in FHIR, ensuring that AI predictions are made based on consistent, normalized data. This makes it possible for even single-message FHIR resources (like a patient’s admission record or a medication order) to be compatible with pre-existing, trained models, which are often designed to analyze more comprehensive patient data typically found in OMOP.
An example of this use case could involve a healthcare provider using an AI model to predict medication adherence for patients. The AI model is trained on historical clinical data stored in the OMOP format, which contains detailed, structured information about patient visits, medications, diagnoses, and more. The healthcare provider collects patient data in real-time via FHIR resources, such as prescriptions and medication refills, which are stored in their EHR system.
Through the ETL process, the FHIR resources (prescription orders, medication administration records, etc.) are transformed into the OMOP CDM format, ensuring compatibility with the trained model. Once transformed, the model can be applied to real-time data to predict whether a patient is likely to adhere to their prescribed medication regimen. The output of the model can be classified based on this prediction and used to guide healthcare interventions.
The FHIR to OMOP transformation enables healthcare organizations to leverage real-time clinical data in FHIR format with pre-trained AI models that rely on the OMOP CDM. Through ETL processes, even fragmented or single-message FHIR resources can be made compatible with AI models trained on more comprehensive datasets, fostering more effective AI-driven healthcare decisions and interventions.