This page is part of the Genetic Reporting Implementation Guide (v3.0.0: STU3) based on FHIR (HL7® FHIR® Standard) R4. This is the current published version in its permanent home (it will always be available at this URL). For a full list of available versions, see the Directory of published versions
| Official URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/implication | Version: 3.0.0 | |||
| Active as of 2024-12-12 | Computable Name: GenomicImplication | |||
Copyright/Legal: This material contains content from LOINC (http://loinc.org). LOINC is copyright © 1995-2020, Regenstrief Institute, Inc. and the Logical Observation Identifiers Names and Codes (LOINC) Committee and is available at no cost under the license at http://loinc.org/license. LOINC® is a registered United States trademark of Regenstrief Institute, Inc. |
||||
Properties common to genomic implications expressed as computable discrete elements.
The Genomic Implication profile is an abstract profile for therapeutic and diagnostic implications and molecular consequences. Implications are relevant information which are derived from the presence of a haplotype, genotype, variant or molecular biomarker. This portion of the implementation guide relies on the content in the General Genomic Reporting and Variant Reporting portions of this implementation guide.
At the heart of many genomic reports are findings (variants, haplotypes, genotypes, biomarkers) and annotations derived from those findings (diagnostic implications, therapeutic implications, predicted molecular consequences), as shown in the following figure:
This profile defines a common set of elements for other Implications:
derivedFrom is a required field that provides an explicit reference from an implication to the variant(s) / haplotype(s) / genotype(s) / molecular biomarker(s) from which the implication is derived.workflow-relatedArtifact (base extension) supports conveying references to citations, supporting documentation and other information relevant to the asserted implication, such as documentation which applies to the full implication Observationworkflow-relatedArtifactComponent (component level extension) supports conveying references to citations, supporting documentation and other information specific to the component's code and value, such as documentation supporting a specific clinical significance or evidence level valueevidence-level (component) indicates the strength of the evidence behind the asserted implicationclinical-significance (component) indicates the clinical impact of an implicationevidence-level: The amount of observed support for the association between an implication and a variant / haplotype / genotype / biomarker. There are dozens if not hundreds of evidence-level value sets, generally providing an ordinal range of codes from low evidence (e.g. 'predicted', 'case report', '1-star') to high evidence (e.g. 'professional society guideline', 'expert panel consensus', '1A').
clinical-significance: The clinical impact of an implication on a person's health. There are dozens if not hundreds of clinical-significance value sets, generally providing an ordinal range of codes from low significance (e.g. 'benign') to high significance (e.g. 'pathogenic', 'oncogenic','predictive of drug response').
While evidence and significance are orthogonal concepts, some codes conflate the two (e.g. 'expert panel consensus of strong efficacy', 'strong evidence refutes the clinical utility of this drug', 'case report evidence of pathogenicity'). In general, using discrete evidence and significance codes allow for a more precise characterization of an implication. Where an implementation is using a conflated code, we recommend communicating that code in the evidence-level field.
Examples of other code sets for evidence-level and clinical-significance:
There are several cases where it is necessary to differentiate 'AND' conditions (e.g. both drug X AND drug Y, in combination, are indicated in the presence of a variant) vs. 'OR' conditions (e.g., either drug X OR drug Y are indicated in the presence of a variant). This situation is not unique to genomic implications, and arises elsewhere within FHIR (e.g. FHIR Search) and outside of FHIR (e.g., ClinVar submission API condition set). To be consistent with other precedents:
Where implications have fields with cardinality > 1, the inclusion of multiple values within a field SHALL indicate an 'AND' condition. An 'OR' condition SHALL be represented by multiple observation instances.
Implication fields affected by this guidance include:
Molecular consequences: derivedFrom, evidence-level, feature-consequence.
Therapeutic implications: derivedFrom, evidence-level, therapeutic-implication, phenotypic-treatment-context, medication-assessed, therapy-assessed.
Diagnostic implications: derivedFrom, evidence-level, predicted-phenotype, genomic-risk-assessment.
Here are examples of these rules with DiagnosticImplication (but the principles would apply to any Implication):
| Use Case | Description | Visual |
|---|---|---|
| Variant X is pathogenic for Disease Y AND Disease Z (in combination) | One DiagnosticImplication with two predicted-phenotype instances for Disease Y and Disease Z that is derivedFrom one Variant for VariantX |
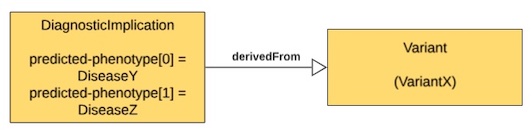 |
| Variant X might be pathogenic for Disease Y alone, or Disease Z alone | Two DiagnosticImplications, one with predicted-phenotype for DiseaseY, second with predicted-phenotype for DiseaseZ, and both are derivedFrom the same Variant for VariantX |
 |
| Variant X AND Variant Y (in combination) are pathogenic for Disease Z | One DiagnosticImplications with predicted-phenotype for DiseaseZ that is derivedFrom the two Variants (VariantX and VariantY) |
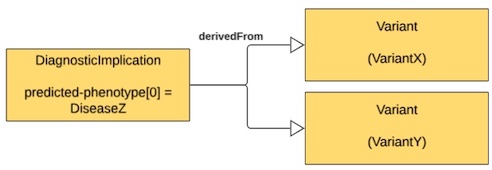 |
| Variant X OR Variant Y might be pathogenic for Disease Z | Two DiagnosticImplications, each with predicted-phenotype for DiseaseZ, one that is derivedFrom a Variant for VariantX, the other that is derivedFrom a Variant for VariantY |
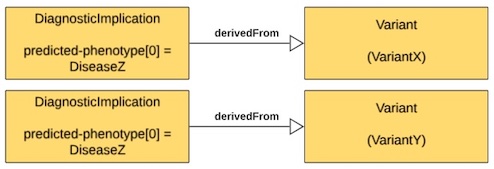 |
Usage:
Description of Profiles, Differentials, Snapshots and how the different presentations work.
This structure is derived from GenomicBase
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 |
0..* | GenomicBase | This is an abstractprofile. Childprofiles: DiagnosticImplication, MolecularConsequence, TherapeuticImplication Measurements and simple assertions | |
  |
Content/Rules for all slices | |||
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |
  |
0..0 | |||
  |
1..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from | |
  |
Content/Rules for all slices | |||
   |
0..* | Reference(Variant) | Variant the implication is derived from | |
   |
0..* | Reference(Genotype) | Genotype the implication is derived from | |
   |
0..* | Reference(Haplotype) | Haplotype the implication is derived from | |
   |
0..* | Reference(Molecular Biomarker) | MolecularBiomarker the implication is derived from | |
  |
0..* | BackboneElement | Component results | |
   |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |
    |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |
  |
Content/Rules for all slices | |||
   |
0..* | BackboneElement | Level of Evidence | |
    |
1..1 | CodeableConcept | 93044-6 Required Pattern: At least the following | |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |
    |
1..1 | CodeableConcept | 1A | 1B | 2A | 2B | 3 | 4 | 4-star | 3-star | 2-star | 1-star | no-star Binding: Evidence Level Examples (example): PharmGKB or ClinVar | |
   |
0..1 | BackboneElement | Clinical significance | |
    |
Content/Rules for all slices | |||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |
    |
1..1 | CodeableConcept | 53037-8 Required Pattern: At least the following | |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 53037-8 | |
    |
1..1 | CodeableConcept | Pathogenic | Likely pathogenic | Uncertain significance | Likely benign | Benign Binding: LOINC Answer List LL4034-6 (example) | |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet | URI |
| Observation.component:evidence-level.value[x] | example | EvidenceLevelExampleVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/evidence-level-example-vsfrom this IG | |
| Observation.component:clinical-significance.value[x] | example | LOINC LL4034-6http://loinc.org/vs/LL4034-6 |
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 |
C | 0..* | GenomicBase | This is an abstractprofile. Childprofiles: DiagnosticImplication, MolecularConsequence, TherapeuticImplication Measurements and simple assertions dom-2: If the resource is contained in another resource, it SHALL NOT contain nested Resources dom-3: If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource dom-4: If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated dom-5: If a resource is contained in another resource, it SHALL NOT have a security label dom-6: A resource should have narrative for robust management obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present |
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created ele-1: All FHIR elements must have a @value or children |
  |
Content/Rules for all slices | |||
   |
0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
   |
0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
  |
?! | 0..* | Extension | Extensions that cannot be ignored ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
  |
?!Σ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. ele-1: All FHIR elements must have a @value or children |
  |
2..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. ele-1: All FHIR elements must have a @value or children | |
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. ele-1: All FHIR elements must have a @value or children | |
    |
Σ | 1..1 | Coding | Code defined by a terminology system ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. ele-1: All FHIR elements must have a @value or children | |
    |
Σ | 1..1 | Coding | Code defined by a terminology system ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |
  |
Σ | 1..1 | CodeableConcept | Type of observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children |
  |
Σ | 1..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from Slice: Unordered, Open by profile:resolve() ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Variant) | Variant the implication is derived from ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Genotype) | Genotype the implication is derived from ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Haplotype) | Haplotype the implication is derived from ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Molecular Biomarker) | MolecularBiomarker the implication is derived from ele-1: All FHIR elements must have a @value or children |
  |
Σ | 0..* | BackboneElement | Component results Slice: Unordered, Open by value:code ele-1: All FHIR elements must have a @value or children |
   |
Content/Rules for all slices | |||
    |
0..* | Extension | Extension Slice: Unordered, Open by value:url ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..1 | BackboneElement | Clinical Conclusion ele-1: All FHIR elements must have a @value or children |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |
   |
Σ | 0..* | BackboneElement | Level of Evidence ele-1: All FHIR elements must have a @value or children |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | 93044-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |
    |
Σ | 1..1 | CodeableConcept | 1A | 1B | 2A | 2B | 3 | 4 | 4-star | 3-star | 2-star | 1-star | no-star Binding: Evidence Level Examples (example): PharmGKB or ClinVar ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..1 | BackboneElement | Clinical significance ele-1: All FHIR elements must have a @value or children |
    |
Content/Rules for all slices | |||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | 53037-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 53037-8 | |
    |
Σ | 1..1 | CodeableConcept | Pathogenic | Likely pathogenic | Uncertain significance | Likely benign | Benign Binding: LOINC Answer List LL4034-6 (example) ele-1: All FHIR elements must have a @value or children |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet / Code | URI |
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | |
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.category:labCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.category:geCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:evidence-level.code | example | Pattern: LOINC Code 93044-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:evidence-level.value[x] | example | EvidenceLevelExampleVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/evidence-level-example-vsfrom this IG | |
| Observation.component:clinical-significance.code | example | Pattern: LOINC Code 53037-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:clinical-significance.value[x] | example | LOINC LL4034-6http://loinc.org/vs/LL4034-6 |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Observation | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Observation | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Observation | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| obs-6 | error | Observation | dataAbsentReason SHALL only be present if Observation.value[x] is not present : dataAbsentReason.empty() or value.empty() | |
| obs-7 | error | Observation | If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present : value.empty() or component.code.where(coding.intersect(%resource.code.coding).exists()).empty() |
| Name | Flags | Card. | Type | Description & Constraints | ||||
|---|---|---|---|---|---|---|---|---|
 |
C | 0..* | GenomicBase | This is an abstractprofile. Childprofiles: DiagnosticImplication, MolecularConsequence, TherapeuticImplication Measurements and simple assertions obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present | ||||
  |
Σ | 0..1 | id | Logical id of this artifact | ||||
  |
Σ | 0..1 | Meta | Metadata about the resource | ||||
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  |
0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  |
0..1 | Narrative | Text summary of the resource, for human interpretation | |||||
  |
0..* | Resource | Contained, inline Resources | |||||
  |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
   |
0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. | |||||
   |
0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |||||
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |||||
  |
?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  |
Σ | 0..* | Identifier | Business Identifier for observation | ||||
  |
Σ | 0..* | Reference(CarePlan | DeviceRequest | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | Fulfills plan, proposal or order | ||||
  |
Σ | 0..* | Reference(MedicationAdministration | MedicationDispense | MedicationStatement | Procedure | Immunization | ImagingStudy | Genomic Study) | Part of referenced event | ||||
  |
?!Σ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. | ||||
  |
2..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |||||
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
     |
0..1 | string | Version of the system - if relevant | |||||
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
     |
0..1 | string | Representation defined by the system | |||||
     |
0..1 | boolean | If this coding was chosen directly by the user | |||||
    |
Σ | 0..1 | string | Plain text representation of the concept | ||||
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |||||
     |
0..1 | string | Version of the system - if relevant | |||||
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |||||
     |
0..1 | string | Representation defined by the system | |||||
     |
0..1 | boolean | If this coding was chosen directly by the user | |||||
    |
Σ | 0..1 | string | Plain text representation of the concept | ||||
  |
Σ | 1..1 | CodeableConcept | Type of observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. | ||||
  |
Σ | 0..1 | Reference(Patient | Group | Device | Location) | Who and/or what the observation is about | ||||
  |
Σ | 0..* | Reference(Resource) | What the observation is about, when it is not about the subject of record | ||||
  |
Σ | 0..1 | Reference(Encounter) | Healthcare event during which this observation is made | ||||
  |
Σ | 0..1 | Clinically relevant time/time-period for observation | |||||
   |
dateTime | |||||||
   |
Period | |||||||
   |
Timing | |||||||
   |
instant | |||||||
  |
Σ | 0..1 | instant | Date/Time this version was made available | ||||
  |
Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam | Patient | RelatedPerson) | Who is responsible for the observation | ||||
  |
C | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
  |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
  |
0..* | CodedAnnotation | Comments about the Observation that also contain a coded type | |||||
  |
0..1 | CodeableConcept | Observed body part Binding: SNOMEDCTBodyStructures (example): Codes describing anatomical locations. May include laterality. | |||||
  |
0..1 | CodeableConcept | How it was done Binding: ObservationMethods (example): Methods for simple observations. | |||||
  |
0..1 | Reference(Specimen) | Specimen used for this observation | |||||
  |
0..1 | Reference(Device | DeviceMetric) | (Measurement) Device | |||||
  |
C | 0..* | BackboneElement | Provides guide for interpretation obs-3: Must have at least a low or a high or text | ||||
   |
0..1 | string | Unique id for inter-element referencing | |||||
   |
0..* | Extension | Additional content defined by implementations | |||||
   |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   |
C | 0..1 | SimpleQuantity | Low Range, if relevant | ||||
   |
C | 0..1 | SimpleQuantity | High Range, if relevant | ||||
   |
0..1 | CodeableConcept | Reference range qualifier Binding: ObservationReferenceRangeMeaningCodes (preferred): Code for the meaning of a reference range. | |||||
   |
0..* | CodeableConcept | Reference range population Binding: ObservationReferenceRangeAppliesToCodes (example): Codes identifying the population the reference range applies to. | |||||
   |
0..1 | Range | Applicable age range, if relevant | |||||
   |
0..1 | string | Text based reference range in an observation | |||||
  |
Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Related resource that belongs to the Observation group | ||||
  |
Σ | 1..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from Slice: Unordered, Open by profile:resolve() | ||||
   |
Σ | 0..* | Reference(Variant) | Variant the implication is derived from | ||||
   |
Σ | 0..* | Reference(Genotype) | Genotype the implication is derived from | ||||
   |
Σ | 0..* | Reference(Haplotype) | Haplotype the implication is derived from | ||||
   |
Σ | 0..* | Reference(Molecular Biomarker) | MolecularBiomarker the implication is derived from | ||||
  |
Σ | 0..* | BackboneElement | Component results Slice: Unordered, Open by value:code | ||||
   |
Content/Rules for all slices | |||||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. | ||||
    |
Σ | 0..1 | Actual component result | |||||
     |
Quantity | |||||||
     |
CodeableConcept | |||||||
     |
string | |||||||
     |
boolean | |||||||
     |
integer | |||||||
     |
Range | |||||||
     |
Ratio | |||||||
     |
SampledData | |||||||
     |
time | |||||||
     |
dateTime | |||||||
     |
Period | |||||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   |
Σ | 0..1 | BackboneElement | Clinical Conclusion | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      |
0..1 | string | Unique id for inter-element referencing | |||||
      |
0..* | Extension | Additional content defined by implementations | |||||
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      |
0..1 | string | Version of the system - if relevant | |||||
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |||||
      |
0..1 | string | Representation defined by the system | |||||
      |
0..1 | boolean | If this coding was chosen directly by the user | |||||
     |
0..1 | string | Plain text representation of the concept | |||||
    |
Σ | 0..1 | string | Summary conclusion (interpretation/impression) | ||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   |
Σ | 0..* | BackboneElement | Level of Evidence | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | 93044-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      |
0..1 | string | Unique id for inter-element referencing | |||||
      |
0..* | Extension | Additional content defined by implementations | |||||
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      |
0..1 | string | Version of the system - if relevant | |||||
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |||||
      |
0..1 | string | Representation defined by the system | |||||
      |
0..1 | boolean | If this coding was chosen directly by the user | |||||
     |
0..1 | string | Plain text representation of the concept | |||||
    |
Σ | 1..1 | CodeableConcept | 1A | 1B | 2A | 2B | 3 | 4 | 4-star | 3-star | 2-star | 1-star | no-star Binding: Evidence Level Examples (example): PharmGKB or ClinVar | ||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   |
Σ | 0..1 | BackboneElement | Clinical significance | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | 53037-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      |
0..1 | string | Unique id for inter-element referencing | |||||
      |
0..* | Extension | Additional content defined by implementations | |||||
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      |
0..1 | string | Version of the system - if relevant | |||||
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 53037-8 | |||||
      |
0..1 | string | Representation defined by the system | |||||
      |
0..1 | boolean | If this coding was chosen directly by the user | |||||
     |
0..1 | string | Plain text representation of the concept | |||||
    |
Σ | 1..1 | CodeableConcept | Pathogenic | Likely pathogenic | Uncertain significance | Likely benign | Benign Binding: LOINC Answer List LL4034-6 (example) | ||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Conformance | ValueSet / Code | URI | |||
| Observation.language | preferred | CommonLanguageshttp://hl7.org/fhir/ValueSet/languagesfrom the FHIR Standard
| ||||
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | ||||
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.category:labCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.category:geCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.bodySite | example | SNOMEDCTBodyStructureshttp://hl7.org/fhir/ValueSet/body-sitefrom the FHIR Standard | ||||
| Observation.method | example | ObservationMethodshttp://hl7.org/fhir/ValueSet/observation-methodsfrom the FHIR Standard | ||||
| Observation.referenceRange.type | preferred | ObservationReferenceRangeMeaningCodeshttp://hl7.org/fhir/ValueSet/referencerange-meaningfrom the FHIR Standard | ||||
| Observation.referenceRange.appliesTo | example | ObservationReferenceRangeAppliesToCodeshttp://hl7.org/fhir/ValueSet/referencerange-appliestofrom the FHIR Standard | ||||
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:evidence-level.code | example | Pattern: LOINC Code 93044-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:evidence-level.value[x] | example | EvidenceLevelExampleVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/evidence-level-example-vsfrom this IG | ||||
| Observation.component:evidence-level.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:evidence-level.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:clinical-significance.code | example | Pattern: LOINC Code 53037-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:clinical-significance.value[x] | example | LOINC LL4034-6http://loinc.org/vs/LL4034-6 | ||||
| Observation.component:clinical-significance.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:clinical-significance.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Observation | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Observation | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Observation | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| obs-3 | error | Observation.referenceRange | Must have at least a low or a high or text : low.exists() or high.exists() or text.exists() | |
| obs-6 | error | Observation | dataAbsentReason SHALL only be present if Observation.value[x] is not present : dataAbsentReason.empty() or value.empty() | |
| obs-7 | error | Observation | If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present : value.empty() or component.code.where(coding.intersect(%resource.code.coding).exists()).empty() |
This structure is derived from GenomicBase
Summary
Mandatory: 1 element(2 nested mandatory elements)
Prohibited: 1 element
Structures
This structure refers to these other structures:
Extensions
This structure refers to these extensions:
Differential View
This structure is derived from GenomicBase
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 |
0..* | GenomicBase | This is an abstractprofile. Childprofiles: DiagnosticImplication, MolecularConsequence, TherapeuticImplication Measurements and simple assertions | |
  |
Content/Rules for all slices | |||
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |
  |
0..0 | |||
  |
1..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from | |
  |
Content/Rules for all slices | |||
   |
0..* | Reference(Variant) | Variant the implication is derived from | |
   |
0..* | Reference(Genotype) | Genotype the implication is derived from | |
   |
0..* | Reference(Haplotype) | Haplotype the implication is derived from | |
   |
0..* | Reference(Molecular Biomarker) | MolecularBiomarker the implication is derived from | |
  |
0..* | BackboneElement | Component results | |
   |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |
    |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |
  |
Content/Rules for all slices | |||
   |
0..* | BackboneElement | Level of Evidence | |
    |
1..1 | CodeableConcept | 93044-6 Required Pattern: At least the following | |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |
    |
1..1 | CodeableConcept | 1A | 1B | 2A | 2B | 3 | 4 | 4-star | 3-star | 2-star | 1-star | no-star Binding: Evidence Level Examples (example): PharmGKB or ClinVar | |
   |
0..1 | BackboneElement | Clinical significance | |
    |
Content/Rules for all slices | |||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |
    |
1..1 | CodeableConcept | 53037-8 Required Pattern: At least the following | |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 53037-8 | |
    |
1..1 | CodeableConcept | Pathogenic | Likely pathogenic | Uncertain significance | Likely benign | Benign Binding: LOINC Answer List LL4034-6 (example) | |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet | URI |
| Observation.component:evidence-level.value[x] | example | EvidenceLevelExampleVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/evidence-level-example-vsfrom this IG | |
| Observation.component:clinical-significance.value[x] | example | LOINC LL4034-6http://loinc.org/vs/LL4034-6 |
Key Elements View
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 |
C | 0..* | GenomicBase | This is an abstractprofile. Childprofiles: DiagnosticImplication, MolecularConsequence, TherapeuticImplication Measurements and simple assertions dom-2: If the resource is contained in another resource, it SHALL NOT contain nested Resources dom-3: If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource dom-4: If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated dom-5: If a resource is contained in another resource, it SHALL NOT have a security label dom-6: A resource should have narrative for robust management obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present |
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created ele-1: All FHIR elements must have a @value or children |
  |
Content/Rules for all slices | |||
   |
0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
   |
0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
  |
?! | 0..* | Extension | Extensions that cannot be ignored ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
  |
?!Σ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. ele-1: All FHIR elements must have a @value or children |
  |
2..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. ele-1: All FHIR elements must have a @value or children | |
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. ele-1: All FHIR elements must have a @value or children | |
    |
Σ | 1..1 | Coding | Code defined by a terminology system ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. ele-1: All FHIR elements must have a @value or children | |
    |
Σ | 1..1 | Coding | Code defined by a terminology system ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |
  |
Σ | 1..1 | CodeableConcept | Type of observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children |
  |
Σ | 1..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from Slice: Unordered, Open by profile:resolve() ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Variant) | Variant the implication is derived from ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Genotype) | Genotype the implication is derived from ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Haplotype) | Haplotype the implication is derived from ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..* | Reference(Molecular Biomarker) | MolecularBiomarker the implication is derived from ele-1: All FHIR elements must have a @value or children |
  |
Σ | 0..* | BackboneElement | Component results Slice: Unordered, Open by value:code ele-1: All FHIR elements must have a @value or children |
   |
Content/Rules for all slices | |||
    |
0..* | Extension | Extension Slice: Unordered, Open by value:url ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..1 | BackboneElement | Clinical Conclusion ele-1: All FHIR elements must have a @value or children |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |
   |
Σ | 0..* | BackboneElement | Level of Evidence ele-1: All FHIR elements must have a @value or children |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | 93044-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |
    |
Σ | 1..1 | CodeableConcept | 1A | 1B | 2A | 2B | 3 | 4 | 4-star | 3-star | 2-star | 1-star | no-star Binding: Evidence Level Examples (example): PharmGKB or ClinVar ele-1: All FHIR elements must have a @value or children |
   |
Σ | 0..1 | BackboneElement | Clinical significance ele-1: All FHIR elements must have a @value or children |
    |
Content/Rules for all slices | |||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both | |
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized ele-1: All FHIR elements must have a @value or children ext-1: Must have either extensions or value[x], not both |
    |
Σ | 1..1 | CodeableConcept | 53037-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. ele-1: All FHIR elements must have a @value or children Required Pattern: At least the following |
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 53037-8 | |
    |
Σ | 1..1 | CodeableConcept | Pathogenic | Likely pathogenic | Uncertain significance | Likely benign | Benign Binding: LOINC Answer List LL4034-6 (example) ele-1: All FHIR elements must have a @value or children |
 Documentation for this format Documentation for this format | ||||
| Path | Conformance | ValueSet / Code | URI |
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | |
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.category:labCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.category:geCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | |
| Observation.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:evidence-level.code | example | Pattern: LOINC Code 93044-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:evidence-level.value[x] | example | EvidenceLevelExampleVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/evidence-level-example-vsfrom this IG | |
| Observation.component:clinical-significance.code | example | Pattern: LOINC Code 53037-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | |
| Observation.component:clinical-significance.value[x] | example | LOINC LL4034-6http://loinc.org/vs/LL4034-6 |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Observation | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Observation | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Observation | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| obs-6 | error | Observation | dataAbsentReason SHALL only be present if Observation.value[x] is not present : dataAbsentReason.empty() or value.empty() | |
| obs-7 | error | Observation | If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present : value.empty() or component.code.where(coding.intersect(%resource.code.coding).exists()).empty() |
Snapshot View
| Name | Flags | Card. | Type | Description & Constraints | ||||
|---|---|---|---|---|---|---|---|---|
 |
C | 0..* | GenomicBase | This is an abstractprofile. Childprofiles: DiagnosticImplication, MolecularConsequence, TherapeuticImplication Measurements and simple assertions obs-6: dataAbsentReason SHALL only be present if Observation.value[x] is not present obs-7: If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present | ||||
  |
Σ | 0..1 | id | Logical id of this artifact | ||||
  |
Σ | 0..1 | Meta | Metadata about the resource | ||||
  |
?!Σ | 0..1 | uri | A set of rules under which this content was created | ||||
  |
0..1 | code | Language of the resource content Binding: CommonLanguages (preferred): A human language.
| |||||
  |
0..1 | Narrative | Text summary of the resource, for human interpretation | |||||
  |
0..* | Resource | Contained, inline Resources | |||||
  |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
   |
0..1 | CodeableConcept | Secondary findings are genetic test results that provide information about variants in a gene unrelated to the primary purpose for the testing, most often discovered when [Whole Exome Sequencing (WES)](https://en.wikipedia.org/wiki/Exome_sequencing) or [Whole Genome Sequencing (WGS)](https://en.wikipedia.org/wiki/Whole_genome_sequencing) is performed. This extension should be used to denote when a genetic finding is being shared as a secondary finding, and ideally refer to a corresponding guideline or policy statement.
For more detail, please see:
https://ghr.nlm.nih.gov/primer/testing/secondaryfindings URL: http://hl7.org/fhir/StructureDefinition/observation-secondaryFinding Binding: GeneticObservationSecondaryFindings (extensible): Codes to denote a guideline or policy statement when a genetic test result is being shared as a secondary finding. | |||||
   |
0..1 | Reference(BodyStructure) | Target anatomic location or structure URL: http://hl7.org/fhir/StructureDefinition/bodySite | |||||
   |
0..* | RelatedArtifact | Documentation relevant to the 'parent' resource URL: http://hl7.org/fhir/StructureDefinition/workflow-relatedArtifact | |||||
  |
?! | 0..* | Extension | Extensions that cannot be ignored | ||||
  |
Σ | 0..* | Identifier | Business Identifier for observation | ||||
  |
Σ | 0..* | Reference(CarePlan | DeviceRequest | ImmunizationRecommendation | MedicationRequest | NutritionOrder | ServiceRequest) | Fulfills plan, proposal or order | ||||
  |
Σ | 0..* | Reference(MedicationAdministration | MedicationDispense | MedicationStatement | Procedure | Immunization | ImagingStudy | Genomic Study) | Part of referenced event | ||||
  |
?!Σ | 1..1 | code | registered | preliminary | final | amended + Binding: ObservationStatus (required): Codes providing the status of an observation. | ||||
  |
2..* | CodeableConcept | Classification of type of observation Slice: Unordered, Open by value:coding Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |||||
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/observation-category | |||||
     |
0..1 | string | Version of the system - if relevant | |||||
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: laboratory | |||||
     |
0..1 | string | Representation defined by the system | |||||
     |
0..1 | boolean | If this coding was chosen directly by the user | |||||
    |
Σ | 0..1 | string | Plain text representation of the concept | ||||
   |
1..1 | CodeableConcept | Classification of type of observation Binding: ObservationCategoryCodes (preferred): Codes for high level observation categories. | |||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations Slice: Unordered, Open by value:url | |||||
    |
Σ | 1..1 | Coding | Code defined by a terminology system Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..1 | uri | Identity of the terminology system Fixed Value: http://terminology.hl7.org/CodeSystem/v2-0074 | |||||
     |
0..1 | string | Version of the system - if relevant | |||||
     |
1..1 | code | Symbol in syntax defined by the system Fixed Value: GE | |||||
     |
0..1 | string | Representation defined by the system | |||||
     |
0..1 | boolean | If this coding was chosen directly by the user | |||||
    |
Σ | 0..1 | string | Plain text representation of the concept | ||||
  |
Σ | 1..1 | CodeableConcept | Type of observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. | ||||
  |
Σ | 0..1 | Reference(Patient | Group | Device | Location) | Who and/or what the observation is about | ||||
  |
Σ | 0..* | Reference(Resource) | What the observation is about, when it is not about the subject of record | ||||
  |
Σ | 0..1 | Reference(Encounter) | Healthcare event during which this observation is made | ||||
  |
Σ | 0..1 | Clinically relevant time/time-period for observation | |||||
   |
dateTime | |||||||
   |
Period | |||||||
   |
Timing | |||||||
   |
instant | |||||||
  |
Σ | 0..1 | instant | Date/Time this version was made available | ||||
  |
Σ | 0..* | Reference(Practitioner | PractitionerRole | Organization | CareTeam | Patient | RelatedPerson) | Who is responsible for the observation | ||||
  |
C | 0..1 | CodeableConcept | Why the result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
  |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
  |
0..* | CodedAnnotation | Comments about the Observation that also contain a coded type | |||||
  |
0..1 | CodeableConcept | Observed body part Binding: SNOMEDCTBodyStructures (example): Codes describing anatomical locations. May include laterality. | |||||
  |
0..1 | CodeableConcept | How it was done Binding: ObservationMethods (example): Methods for simple observations. | |||||
  |
0..1 | Reference(Specimen) | Specimen used for this observation | |||||
  |
0..1 | Reference(Device | DeviceMetric) | (Measurement) Device | |||||
  |
C | 0..* | BackboneElement | Provides guide for interpretation obs-3: Must have at least a low or a high or text | ||||
   |
0..1 | string | Unique id for inter-element referencing | |||||
   |
0..* | Extension | Additional content defined by implementations | |||||
   |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
   |
C | 0..1 | SimpleQuantity | Low Range, if relevant | ||||
   |
C | 0..1 | SimpleQuantity | High Range, if relevant | ||||
   |
0..1 | CodeableConcept | Reference range qualifier Binding: ObservationReferenceRangeMeaningCodes (preferred): Code for the meaning of a reference range. | |||||
   |
0..* | CodeableConcept | Reference range population Binding: ObservationReferenceRangeAppliesToCodes (example): Codes identifying the population the reference range applies to. | |||||
   |
0..1 | Range | Applicable age range, if relevant | |||||
   |
0..1 | string | Text based reference range in an observation | |||||
  |
Σ | 0..* | Reference(Observation | QuestionnaireResponse | MolecularSequence) | Related resource that belongs to the Observation group | ||||
  |
Σ | 1..* | Reference(DocumentReference | ImagingStudy | Media | QuestionnaireResponse | Observation | MolecularSequence) | Related measurements the observation is made from Slice: Unordered, Open by profile:resolve() | ||||
   |
Σ | 0..* | Reference(Variant) | Variant the implication is derived from | ||||
   |
Σ | 0..* | Reference(Genotype) | Genotype the implication is derived from | ||||
   |
Σ | 0..* | Reference(Haplotype) | Haplotype the implication is derived from | ||||
   |
Σ | 0..* | Reference(Molecular Biomarker) | MolecularBiomarker the implication is derived from | ||||
  |
Σ | 0..* | BackboneElement | Component results Slice: Unordered, Open by value:code | ||||
   |
Content/Rules for all slices | |||||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | Type of component observation (code / type) Binding: LOINCCodes (example): Codes identifying names of simple observations. | ||||
    |
Σ | 0..1 | Actual component result | |||||
     |
Quantity | |||||||
     |
CodeableConcept | |||||||
     |
string | |||||||
     |
boolean | |||||||
     |
integer | |||||||
     |
Range | |||||||
     |
Ratio | |||||||
     |
SampledData | |||||||
     |
time | |||||||
     |
dateTime | |||||||
     |
Period | |||||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   |
Σ | 0..1 | BackboneElement | Clinical Conclusion | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | conclusion-string Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      |
0..1 | string | Unique id for inter-element referencing | |||||
      |
0..* | Extension | Additional content defined by implementations | |||||
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://hl7.org/fhir/uv/genomics-reporting/CodeSystem/tbd-codes-cs | |||||
      |
0..1 | string | Version of the system - if relevant | |||||
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: conclusion-string | |||||
      |
0..1 | string | Representation defined by the system | |||||
      |
0..1 | boolean | If this coding was chosen directly by the user | |||||
     |
0..1 | string | Plain text representation of the concept | |||||
    |
Σ | 0..1 | string | Summary conclusion (interpretation/impression) | ||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   |
Σ | 0..* | BackboneElement | Level of Evidence | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Additional content defined by implementations | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | 93044-6 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      |
0..1 | string | Unique id for inter-element referencing | |||||
      |
0..* | Extension | Additional content defined by implementations | |||||
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      |
0..1 | string | Version of the system - if relevant | |||||
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 93044-6 | |||||
      |
0..1 | string | Representation defined by the system | |||||
      |
0..1 | boolean | If this coding was chosen directly by the user | |||||
     |
0..1 | string | Plain text representation of the concept | |||||
    |
Σ | 1..1 | CodeableConcept | 1A | 1B | 2A | 2B | 3 | 4 | 4-star | 3-star | 2-star | 1-star | no-star Binding: Evidence Level Examples (example): PharmGKB or ClinVar | ||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
   |
Σ | 0..1 | BackboneElement | Clinical significance | ||||
    |
0..1 | string | Unique id for inter-element referencing | |||||
    |
0..* | Extension | Extension Slice: Unordered, Open by value:url | |||||
     |
0..* | RelatedArtifact | Related Artifact for Observation component URL: http://hl7.org/fhir/uv/genomics-reporting/StructureDefinition/workflow-relatedArtifactComponent | |||||
    |
?!Σ | 0..* | Extension | Extensions that cannot be ignored even if unrecognized | ||||
    |
Σ | 1..1 | CodeableConcept | 53037-8 Binding: LOINCCodes (example): Codes identifying names of simple observations. Required Pattern: At least the following | ||||
     |
0..1 | string | Unique id for inter-element referencing | |||||
     |
0..* | Extension | Additional content defined by implementations | |||||
     |
1..* | Coding | Code defined by a terminology system Fixed Value: (complex) | |||||
      |
0..1 | string | Unique id for inter-element referencing | |||||
      |
0..* | Extension | Additional content defined by implementations | |||||
      |
1..1 | uri | Identity of the terminology system Fixed Value: http://loinc.org | |||||
      |
0..1 | string | Version of the system - if relevant | |||||
      |
1..1 | code | Symbol in syntax defined by the system Fixed Value: 53037-8 | |||||
      |
0..1 | string | Representation defined by the system | |||||
      |
0..1 | boolean | If this coding was chosen directly by the user | |||||
     |
0..1 | string | Plain text representation of the concept | |||||
    |
Σ | 1..1 | CodeableConcept | Pathogenic | Likely pathogenic | Uncertain significance | Likely benign | Benign Binding: LOINC Answer List LL4034-6 (example) | ||||
    |
C | 0..1 | CodeableConcept | Why the component result is missing Binding: DataAbsentReason (extensible): Codes specifying why the result (Observation.value[x]) is missing. | ||||
    |
0..* | CodeableConcept | High, low, normal, etc. Binding: ObservationInterpretationCodes (extensible): Codes identifying interpretations of observations. | |||||
    |
0..* | See referenceRange (Observation) | Provides guide for interpretation of component result | |||||
 Documentation for this format Documentation for this format | ||||||||
| Path | Conformance | ValueSet / Code | URI | |||
| Observation.language | preferred | CommonLanguageshttp://hl7.org/fhir/ValueSet/languagesfrom the FHIR Standard
| ||||
| Observation.status | required | ObservationStatushttp://hl7.org/fhir/ValueSet/observation-status|4.0.1from the FHIR Standard | ||||
| Observation.category | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.category:labCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.category:geCategory | preferred | ObservationCategoryCodeshttp://hl7.org/fhir/ValueSet/observation-categoryfrom the FHIR Standard | ||||
| Observation.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.bodySite | example | SNOMEDCTBodyStructureshttp://hl7.org/fhir/ValueSet/body-sitefrom the FHIR Standard | ||||
| Observation.method | example | ObservationMethodshttp://hl7.org/fhir/ValueSet/observation-methodsfrom the FHIR Standard | ||||
| Observation.referenceRange.type | preferred | ObservationReferenceRangeMeaningCodeshttp://hl7.org/fhir/ValueSet/referencerange-meaningfrom the FHIR Standard | ||||
| Observation.referenceRange.appliesTo | example | ObservationReferenceRangeAppliesToCodeshttp://hl7.org/fhir/ValueSet/referencerange-appliestofrom the FHIR Standard | ||||
| Observation.component.code | example | LOINCCodes (a valid code from LOINC)http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.code | example | Pattern: conclusion-stringhttp://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:conclusion-string.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:evidence-level.code | example | Pattern: LOINC Code 93044-6http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:evidence-level.value[x] | example | EvidenceLevelExampleVShttp://hl7.org/fhir/uv/genomics-reporting/ValueSet/evidence-level-example-vsfrom this IG | ||||
| Observation.component:evidence-level.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:evidence-level.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard | ||||
| Observation.component:clinical-significance.code | example | Pattern: LOINC Code 53037-8http://hl7.org/fhir/ValueSet/observation-codesfrom the FHIR Standard | ||||
| Observation.component:clinical-significance.value[x] | example | LOINC LL4034-6http://loinc.org/vs/LL4034-6 | ||||
| Observation.component:clinical-significance.dataAbsentReason | extensible | DataAbsentReasonhttp://hl7.org/fhir/ValueSet/data-absent-reasonfrom the FHIR Standard | ||||
| Observation.component:clinical-significance.interpretation | extensible | ObservationInterpretationCodeshttp://hl7.org/fhir/ValueSet/observation-interpretationfrom the FHIR Standard |
| Id | Grade | Path(s) | Details | Requirements |
| dom-2 | error | Observation | If the resource is contained in another resource, it SHALL NOT contain nested Resources : contained.contained.empty() | |
| dom-3 | error | Observation | If the resource is contained in another resource, it SHALL be referred to from elsewhere in the resource or SHALL refer to the containing resource : contained.where((('#'+id in (%resource.descendants().reference | %resource.descendants().as(canonical) | %resource.descendants().as(uri) | %resource.descendants().as(url))) or descendants().where(reference = '#').exists() or descendants().where(as(canonical) = '#').exists() or descendants().where(as(canonical) = '#').exists()).not()).trace('unmatched', id).empty() | |
| dom-4 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a meta.versionId or a meta.lastUpdated : contained.meta.versionId.empty() and contained.meta.lastUpdated.empty() | |
| dom-5 | error | Observation | If a resource is contained in another resource, it SHALL NOT have a security label : contained.meta.security.empty() | |
| dom-6 | best practice | Observation | A resource should have narrative for robust management : text.`div`.exists() | |
| ele-1 | error | **ALL** elements | All FHIR elements must have a @value or children : hasValue() or (children().count() > id.count()) | |
| ext-1 | error | **ALL** extensions | Must have either extensions or value[x], not both : extension.exists() != value.exists() | |
| obs-3 | error | Observation.referenceRange | Must have at least a low or a high or text : low.exists() or high.exists() or text.exists() | |
| obs-6 | error | Observation | dataAbsentReason SHALL only be present if Observation.value[x] is not present : dataAbsentReason.empty() or value.empty() | |
| obs-7 | error | Observation | If Observation.code is the same as an Observation.component.code then the value element associated with the code SHALL NOT be present : value.empty() or component.code.where(coding.intersect(%resource.code.coding).exists()).empty() |
This structure is derived from GenomicBase
Summary
Mandatory: 1 element(2 nested mandatory elements)
Prohibited: 1 element
Structures
This structure refers to these other structures:
Extensions
This structure refers to these extensions:
Other representations of profile: CSV, Excel, Schematron