This page is part of the FHIR Specification (v5.0.0-draft-final: Final QA Preview for R5 - see ballot notes). The current version which supercedes this version is 5.0.0. For a full list of available versions, see the Directory of published versions 
Clinical Genomics  Work Group Work Group | Maturity Level: 0 | Trial Use | Security Category: Patient | Compartments: Patient |
A Genomic Study is a set of analysis performed to analyze and generate genomic data.
Note to Implementers: The material on this page is currently undergoing work to be refactored in a future release as further analysis is done. Reviewers are encouraged to provide their comments and feedback about the example codes for terminology binding purposes.
GenomicStudy resource aims at delineating relevant information of a genomic study. A genomic study might comprise one or more analyses, each serving a specific purpose. These analyses may vary in method (e.g., karyotyping, CNV, or SNV detection), performer, software, devices used, or regions targeted.
GenomicStudy describes relevant information including the reasons, purpose, and performers of the study. It also provides technical endpoints to access these data. It could be a logical aggregator for complex genomic analyses.
A genomic study might be based on one or more service requests or care plans. The reasons for performing a genomic study might be driven by specific Conditions or Observations. Although the genomic study subject is the focus of the study, the analysis-level focus could be used to specify other relevant subjects or specimens. For example, trio testing may involve three subjects: the proband, and both parents. The proband would be the subject listed directly under the genomic study, while each of the analyses could represent proband, mother, and father genomic analyses.
In clinical use, the study interpreters use all the results of these various analyses to provide diagnostic and therapeutic insights for the patient. Where performers are listed according to their participation in each genomic analysis. Each analysis may be based on a set of defined protocols that may differ from the performed protocols.
The regionStudied and regionCalled elements can refer to DocumentReference instances carrying BED files. Other types of files used in an analysis can be provided similarly, and listed either as inputs or outputs.
Detailed information about the results of the analyses may be represented by Observations and gathered in a DiagnosticReport. Both resources may refer back to the originating GenomicStudy to allow navigation.
A main function of GenomicStudy is to provide additional metadata for one or more clinical genomic analysis pipelines where DocumentReference instances are used to track inputs and outputs of those pipelines. Device is used to represent both hardware and software used in the sequencing and in the analyses.
It is intended that this resource be referenced by Observation and DiagnosticReport similarly to how ImagingStudy is currently referenced on those resources. Of note, the results of the study would be captured in Observations and referenced on a DiagnosticReport.
Important contextual references include Patient, Specimen, Encounter, ServiceRequest, Practitioner, and Organization.
Observation or Condition instances may also be referenced from genomic study to designate a given reason for performing the study.
GenomicStudy can support the event-definition workflow pattern by referencing PlanDefinition via the instantiatesCanonical relationship when implementers wish to describe more specifics of the pipelines and devices used in the analysis.
Structure
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | TU | DomainResource | Genomic Study Elements defined in Ancestors: id, meta, implicitRules, language, text, contained, extension, modifierExtension | |
  | Σ | 0..* | Identifier | Identifiers for this genomic study |
  | ?!Σ | 1..1 | code | registered | available | cancelled | entered-in-error | unknown Binding: Genomic Study Status (Required) |
  | Σ | 0..* | CodeableConcept | The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling) Binding: Genomic Study Type (Example) |
  | Σ | 1..1 | Reference(Patient | Group | Substance | BiologicallyDerivedProduct | NutritionProduct) | The primary subject of the genomic study |
  | Σ | 0..1 | Reference(Encounter) | The healthcare event with which this genomics study is associated |
  | 0..1 | dateTime | When the genomic study was started | |
  | 0..* | Reference(ServiceRequest | Task) | Event resources that the genomic study is based on | |
  | 0..1 | Reference(Practitioner | PractitionerRole) | Healthcare professional who requested or referred the genomic study | |
  | 0..* | Reference(Practitioner | PractitionerRole) | Healthcare professionals who interpreted the genomic study | |
  | 0..* | CodeableReference(Condition | Observation) | Why the genomic study was performed | |
  | 0..1 | canonical(PlanDefinition) | The defined protocol that describes the study | |
  | 0..1 | uri | The URL pointing to an externally maintained protocol that describes the study | |
  | 0..* | Annotation | Comments related to the genomic study | |
  | 0..1 | markdown | Description of the genomic study | |
  | 0..* | BackboneElement | Genomic Analysis Event | |
   | Σ | 0..* | Identifier | Identifiers for the analysis event |
   | Σ | 0..* | CodeableConcept | Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI) Binding: Genomic Study Method Type (Example) |
   | 0..* | CodeableConcept | Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change) Binding: Genomic Study Change Type (Example) | |
   | 0..1 | CodeableConcept | Genome build that is used in this analysis Binding: HumanRefSeqNCBIBuildId  (Extensible) (Extensible) | |
   | 0..1 | canonical(PlanDefinition | ActivityDefinition) | The defined protocol that describes the analysis | |
   | 0..1 | uri | The URL pointing to an externally maintained protocol that describes the analysis | |
   | Σ | 0..1 | string | Name of the analysis event (human friendly) |
   | Σ | 0..* | Reference(Any) | What the genomic analysis is about, when it is not about the subject of record |
   | Σ | 0..* | Reference(Specimen) | The specimen used in the analysis event |
   | 0..1 | dateTime | The date of the analysis event | |
   | 0..* | Annotation | Any notes capture with the analysis event | |
   | 0..1 | Reference(Procedure | Task) | The protocol that was performed for the analysis event | |
   | 0..* | Reference(DocumentReference | Observation) | The genomic regions to be studied in the analysis (BED file) | |
   | 0..* | Reference(DocumentReference | Observation) | Genomic regions actually called in the analysis event (BED file) | |
   | 0..* | BackboneElement | Inputs for the analysis event | |
  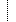  | Σ | 0..1 | Reference(DocumentReference) | File containing input data |
  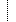  | 0..1 | CodeableConcept | Type of input data (e.g., BAM, CRAM, or FASTA) Binding: Genomic Study Data Format (Example) | |
  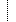  | 0..1 | The analysis event or other GenomicStudy that generated this input file | ||
  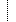   | Identifier | |||
  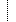  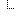 | Reference(GenomicStudy) | |||
   | 0..* | BackboneElement | Outputs for the analysis event | |
  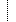  | Σ | 0..1 | Reference(DocumentReference) | File containing output data |
  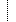  | Σ | 0..1 | CodeableConcept | Type of output data (e.g., VCF, MAF, or BAM) Binding: Genomic Study Data Format (Example) |
   | 0..* | BackboneElement | Performer for the analysis event | |
    | 0..1 | Reference(Practitioner | PractitionerRole | Organization | Device) | The organization, healthcare professional, or others who participated in performing this analysis | |
    | 0..1 | CodeableConcept | Role of the actor for this analysis | |
   | 0..* | BackboneElement | Devices used for the analysis (e.g., instruments, software), with settings and parameters | |
    | 0..1 | Reference(Device) | Device used for the analysis | |
    | 0..1 | CodeableConcept | Specific function for the device used for the analysis | |
 Documentation for this format Documentation for this format  | ||||
See the Extensions for this resource
UML Diagram (Legend)
XML Template
<GenomicStudy xmlns="http://hl7.org/fhir"><!-- from Resource: id, meta, implicitRules, and language --> <!-- from DomainResource: text, contained, extension, and modifierExtension --> <identifier><!-- 0..* Identifier Identifiers for this genomic study --></identifier> <status value="[code]"/><!-- 1..1 registered | available | cancelled | entered-in-error | unknown --> <type><!-- 0..* CodeableConcept The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling) --></type> <subject><!-- 1..1 Reference(BiologicallyDerivedProduct|Group|NutritionProduct| Patient|Substance) The primary subject of the genomic study --></subject> <encounter><!-- 0..1 Reference(Encounter) The healthcare event with which this genomics study is associated --></encounter> <startDate value="[dateTime]"/><!-- 0..1 When the genomic study was started --> <basedOn><!-- 0..* Reference(ServiceRequest|Task) Event resources that the genomic study is based on --></basedOn> <referrer><!-- 0..1 Reference(Practitioner|PractitionerRole) Healthcare professional who requested or referred the genomic study --></referrer> <interpreter><!-- 0..* Reference(Practitioner|PractitionerRole) Healthcare professionals who interpreted the genomic study --></interpreter> <reason><!-- 0..* CodeableReference(Condition|Observation) Why the genomic study was performed --></reason> <instantiatesCanonical><!-- 0..1 canonical(PlanDefinition) The defined protocol that describes the study --></instantiatesCanonical> <instantiatesUri value="[uri]"/><!-- 0..1 The URL pointing to an externally maintained protocol that describes the study --> <note><!-- 0..* Annotation Comments related to the genomic study --></note> <description value="[markdown]"/><!-- 0..1 Description of the genomic study --> <analysis> <!-- 0..* Genomic Analysis Event --> <identifier><!-- 0..* Identifier Identifiers for the analysis event --></identifier> <methodType><!-- 0..* CodeableConcept Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI) --></methodType> <changeType><!-- 0..* CodeableConcept Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change) --></changeType> <genomeBuild><!-- 0..1 CodeableConcept Genome build that is used in this analysis
--></genomeBuild> <instantiatesCanonical><!-- 0..1 canonical(ActivityDefinition|PlanDefinition) The defined protocol that describes the analysis --></instantiatesCanonical> <instantiatesUri value="[uri]"/><!-- 0..1 The URL pointing to an externally maintained protocol that describes the analysis --> <title value="[string]"/><!-- 0..1 Name of the analysis event (human friendly) --> <focus><!-- 0..* Reference(Any) What the genomic analysis is about, when it is not about the subject of record --></focus> <specimen><!-- 0..* Reference(Specimen) The specimen used in the analysis event --></specimen> <date value="[dateTime]"/><!-- 0..1 The date of the analysis event --> <note><!-- 0..* Annotation Any notes capture with the analysis event --></note> <protocolPerformed><!-- 0..1 Reference(Procedure|Task) The protocol that was performed for the analysis event --></protocolPerformed> <regionsStudied><!-- 0..* Reference(DocumentReference|Observation) The genomic regions to be studied in the analysis (BED file) --></regionsStudied> <regionsCalled><!-- 0..* Reference(DocumentReference|Observation) Genomic regions actually called in the analysis event (BED file) --></regionsCalled> <input> <!-- 0..* Inputs for the analysis event --> <file><!-- 0..1 Reference(DocumentReference) File containing input data --></file> <type><!-- 0..1 CodeableConcept Type of input data (e.g., BAM, CRAM, or FASTA) --></type> <generatedBy[x]><!-- 0..1 Identifier|Reference(GenomicStudy) The analysis event or other GenomicStudy that generated this input file --></generatedBy[x]> </input> <output> <!-- 0..* Outputs for the analysis event --> <file><!-- 0..1 Reference(DocumentReference) File containing output data --></file> <type><!-- 0..1 CodeableConcept Type of output data (e.g., VCF, MAF, or BAM) --></type> </output> <performer> <!-- 0..* Performer for the analysis event --> <actor><!-- 0..1 Reference(Device|Organization|Practitioner|PractitionerRole) The organization, healthcare professional, or others who participated in performing this analysis --></actor> <role><!-- 0..1 CodeableConcept Role of the actor for this analysis --></role> </performer> <device> <!-- 0..* Devices used for the analysis (e.g., instruments, software), with settings and parameters --> <device><!-- 0..1 Reference(Device) Device used for the analysis --></device> <function><!-- 0..1 CodeableConcept Specific function for the device used for the analysis --></function> </device> </analysis> </GenomicStudy>
JSON Template
{ "resourceType" : "GenomicStudy",
// from Resource: id, meta, implicitRules, and language
// from DomainResource: text, contained, extension, and modifierExtension
"identifier" : [{ Identifier }], // Identifiers for this genomic study
"status" : "<code>", // R! registered | available | cancelled | entered-in-error | unknown
"type" : [{ CodeableConcept }], // The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling)
"subject" : { Reference(BiologicallyDerivedProduct|Group|NutritionProduct|
Patient|Substance) }, // R! The primary subject of the genomic study
"encounter" : { Reference(Encounter) }, // The healthcare event with which this genomics study is associated
"startDate" : "<dateTime>", // When the genomic study was started
"basedOn" : [{ Reference(ServiceRequest|Task) }], // Event resources that the genomic study is based on
"referrer" : { Reference(Practitioner|PractitionerRole) }, // Healthcare professional who requested or referred the genomic study
"interpreter" : [{ Reference(Practitioner|PractitionerRole) }], // Healthcare professionals who interpreted the genomic study
"reason" : [{ CodeableReference(Condition|Observation) }], // Why the genomic study was performed
"instantiatesCanonical" : "<canonical(PlanDefinition)>", // The defined protocol that describes the study
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the study
"note" : [{ Annotation }], // Comments related to the genomic study
"description" : "<markdown>", // Description of the genomic study
"analysis" : [{ // Genomic Analysis Event
"identifier" : [{ Identifier }], // Identifiers for the analysis event
"methodType" : [{ CodeableConcept }], // Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI)
"changeType" : [{ CodeableConcept }], // Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change)
"genomeBuild" : { CodeableConcept }, // Genome build that is used in this analysis
"resourceType" : "GenomicStudy",
// from Resource: id, meta, implicitRules, and language
// from DomainResource: text, contained, extension, and modifierExtension
"identifier" : [{ Identifier }], // Identifiers for this genomic study
"status" : "<code>", // R! registered | available | cancelled | entered-in-error | unknown
"type" : [{ CodeableConcept }], // The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling)
"subject" : { Reference(BiologicallyDerivedProduct|Group|NutritionProduct|
Patient|Substance) }, // R! The primary subject of the genomic study
"encounter" : { Reference(Encounter) }, // The healthcare event with which this genomics study is associated
"startDate" : "<dateTime>", // When the genomic study was started
"basedOn" : [{ Reference(ServiceRequest|Task) }], // Event resources that the genomic study is based on
"referrer" : { Reference(Practitioner|PractitionerRole) }, // Healthcare professional who requested or referred the genomic study
"interpreter" : [{ Reference(Practitioner|PractitionerRole) }], // Healthcare professionals who interpreted the genomic study
"reason" : [{ CodeableReference(Condition|Observation) }], // Why the genomic study was performed
"instantiatesCanonical" : "<canonical(PlanDefinition)>", // The defined protocol that describes the study
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the study
"note" : [{ Annotation }], // Comments related to the genomic study
"description" : "<markdown>", // Description of the genomic study
"analysis" : [{ // Genomic Analysis Event
"identifier" : [{ Identifier }], // Identifiers for the analysis event
"methodType" : [{ CodeableConcept }], // Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI)
"changeType" : [{ CodeableConcept }], // Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change)
"genomeBuild" : { CodeableConcept }, // Genome build that is used in this analysis  "instantiatesCanonical" : "<canonical(PlanDefinition|ActivityDefinition)>", // The defined protocol that describes the analysis
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the analysis
"title" : "<string>", // Name of the analysis event (human friendly)
"focus" : [{ Reference(Any) }], // What the genomic analysis is about, when it is not about the subject of record
"specimen" : [{ Reference(Specimen) }], // The specimen used in the analysis event
"date" : "<dateTime>", // The date of the analysis event
"note" : [{ Annotation }], // Any notes capture with the analysis event
"protocolPerformed" : { Reference(Procedure|Task) }, // The protocol that was performed for the analysis event
"regionsStudied" : [{ Reference(DocumentReference|Observation) }], // The genomic regions to be studied in the analysis (BED file)
"regionsCalled" : [{ Reference(DocumentReference|Observation) }], // Genomic regions actually called in the analysis event (BED file)
"input" : [{ // Inputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing input data
"type" : { CodeableConcept }, // Type of input data (e.g., BAM, CRAM, or FASTA)
// generatedBy[x]: The analysis event or other GenomicStudy that generated this input file. One of these 2:
"generatedByIdentifier" : { Identifier },
"generatedByReference" : { Reference(GenomicStudy) }
}],
"output" : [{ // Outputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing output data
"type" : { CodeableConcept } // Type of output data (e.g., VCF, MAF, or BAM)
}],
"performer" : [{ // Performer for the analysis event
"actor" : { Reference(Device|Organization|Practitioner|PractitionerRole) }, // The organization, healthcare professional, or others who participated in performing this analysis
"role" : { CodeableConcept } // Role of the actor for this analysis
}],
"device" : [{ // Devices used for the analysis (e.g., instruments, software), with settings and parameters
"device" : { Reference(Device) }, // Device used for the analysis
"function" : { CodeableConcept } // Specific function for the device used for the analysis
}]
}]
}
"instantiatesCanonical" : "<canonical(PlanDefinition|ActivityDefinition)>", // The defined protocol that describes the analysis
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the analysis
"title" : "<string>", // Name of the analysis event (human friendly)
"focus" : [{ Reference(Any) }], // What the genomic analysis is about, when it is not about the subject of record
"specimen" : [{ Reference(Specimen) }], // The specimen used in the analysis event
"date" : "<dateTime>", // The date of the analysis event
"note" : [{ Annotation }], // Any notes capture with the analysis event
"protocolPerformed" : { Reference(Procedure|Task) }, // The protocol that was performed for the analysis event
"regionsStudied" : [{ Reference(DocumentReference|Observation) }], // The genomic regions to be studied in the analysis (BED file)
"regionsCalled" : [{ Reference(DocumentReference|Observation) }], // Genomic regions actually called in the analysis event (BED file)
"input" : [{ // Inputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing input data
"type" : { CodeableConcept }, // Type of input data (e.g., BAM, CRAM, or FASTA)
// generatedBy[x]: The analysis event or other GenomicStudy that generated this input file. One of these 2:
"generatedByIdentifier" : { Identifier },
"generatedByReference" : { Reference(GenomicStudy) }
}],
"output" : [{ // Outputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing output data
"type" : { CodeableConcept } // Type of output data (e.g., VCF, MAF, or BAM)
}],
"performer" : [{ // Performer for the analysis event
"actor" : { Reference(Device|Organization|Practitioner|PractitionerRole) }, // The organization, healthcare professional, or others who participated in performing this analysis
"role" : { CodeableConcept } // Role of the actor for this analysis
}],
"device" : [{ // Devices used for the analysis (e.g., instruments, software), with settings and parameters
"device" : { Reference(Device) }, // Device used for the analysis
"function" : { CodeableConcept } // Specific function for the device used for the analysis
}]
}]
}
Turtle Template
@prefix fhir: <http://hl7.org/fhir/> .[ a fhir:GenomicStudy; fhir:nodeRole fhir:treeRoot; # if this is the parser root # from Resource: .id, .meta, .implicitRules, and .language # from DomainResource: .text, .contained, .extension, and .modifierExtension fhir:identifier ( [ Identifier ] ... ) ; # 0..* Identifiers for this genomic study fhir:status [ code ] ; # 1..1 registered | available | cancelled | entered-in-error | unknown fhir:type ( [ CodeableConcept ] ... ) ; # 0..* The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling) fhir:subject [ Reference(BiologicallyDerivedProduct|Group|NutritionProduct|Patient|Substance) ] ; # 1..1 The primary subject of the genomic study fhir:encounter [ Reference(Encounter) ] ; # 0..1 The healthcare event with which this genomics study is associated fhir:startDate [ dateTime ] ; # 0..1 When the genomic study was started fhir:basedOn ( [ Reference(ServiceRequest|Task) ] ... ) ; # 0..* Event resources that the genomic study is based on fhir:referrer [ Reference(Practitioner|PractitionerRole) ] ; # 0..1 Healthcare professional who requested or referred the genomic study fhir:interpreter ( [ Reference(Practitioner|PractitionerRole) ] ... ) ; # 0..* Healthcare professionals who interpreted the genomic study fhir:reason ( [ CodeableReference(Condition|Observation) ] ... ) ; # 0..* Why the genomic study was performed fhir:instantiatesCanonical [ canonical(PlanDefinition) ] ; # 0..1 The defined protocol that describes the study fhir:instantiatesUri [ uri ] ; # 0..1 The URL pointing to an externally maintained protocol that describes the study fhir:note ( [ Annotation ] ... ) ; # 0..* Comments related to the genomic study fhir:description [ markdown ] ; # 0..1 Description of the genomic study fhir:analysis ( [ # 0..* Genomic Analysis Event fhir:identifier ( [ Identifier ] ... ) ; # 0..* Identifiers for the analysis event fhir:methodType ( [ CodeableConcept ] ... ) ; # 0..* Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI) fhir:changeType ( [ CodeableConcept ] ... ) ; # 0..* Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change) fhir:genomeBuild [ CodeableConcept ] ; # 0..1 Genome build that is used in this analysis fhir:instantiatesCanonical [ canonical(ActivityDefinition|PlanDefinition) ] ; # 0..1 The defined protocol that describes the analysis fhir:instantiatesUri [ uri ] ; # 0..1 The URL pointing to an externally maintained protocol that describes the analysis fhir:title [ string ] ; # 0..1 Name of the analysis event (human friendly) fhir:focus ( [ Reference(Any) ] ... ) ; # 0..* What the genomic analysis is about, when it is not about the subject of record fhir:specimen ( [ Reference(Specimen) ] ... ) ; # 0..* The specimen used in the analysis event fhir:date [ dateTime ] ; # 0..1 The date of the analysis event fhir:note ( [ Annotation ] ... ) ; # 0..* Any notes capture with the analysis event fhir:protocolPerformed [ Reference(Procedure|Task) ] ; # 0..1 The protocol that was performed for the analysis event fhir:regionsStudied ( [ Reference(DocumentReference|Observation) ] ... ) ; # 0..* The genomic regions to be studied in the analysis (BED file) fhir:regionsCalled ( [ Reference(DocumentReference|Observation) ] ... ) ; # 0..* Genomic regions actually called in the analysis event (BED file) fhir:input ( [ # 0..* Inputs for the analysis event fhir:file [ Reference(DocumentReference) ] ; # 0..1 File containing input data fhir:type [ CodeableConcept ] ; # 0..1 Type of input data (e.g., BAM, CRAM, or FASTA) # generatedBy[x] : 0..1 The analysis event or other GenomicStudy that generated this input file. One of these 2 fhir:generatedBy [ a fhir:Identifier ; Identifier ] fhir:generatedBy [ a fhir:Reference ; Reference(GenomicStudy) ] ] ... ) ; fhir:output ( [ # 0..* Outputs for the analysis event fhir:file [ Reference(DocumentReference) ] ; # 0..1 File containing output data fhir:type [ CodeableConcept ] ; # 0..1 Type of output data (e.g., VCF, MAF, or BAM) ] ... ) ; fhir:performer ( [ # 0..* Performer for the analysis event fhir:actor [ Reference(Device|Organization|Practitioner|PractitionerRole) ] ; # 0..1 The organization, healthcare professional, or others who participated in performing this analysis fhir:role [ CodeableConcept ] ; # 0..1 Role of the actor for this analysis ] ... ) ; fhir:device ( [ # 0..* Devices used for the analysis (e.g., instruments, software), with settings and parameters fhir:device [ Reference(Device) ] ; # 0..1 Device used for the analysis fhir:function [ CodeableConcept ] ; # 0..1 Specific function for the device used for the analysis ] ... ) ; ] ... ) ; ]
Structure
| Name | Flags | Card. | Type | Description & Constraints |
|---|---|---|---|---|
 | TU | DomainResource | Genomic Study Elements defined in Ancestors: id, meta, implicitRules, language, text, contained, extension, modifierExtension | |
  | Σ | 0..* | Identifier | Identifiers for this genomic study |
  | ?!Σ | 1..1 | code | registered | available | cancelled | entered-in-error | unknown Binding: Genomic Study Status (Required) |
  | Σ | 0..* | CodeableConcept | The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling) Binding: Genomic Study Type (Example) |
  | Σ | 1..1 | Reference(Patient | Group | Substance | BiologicallyDerivedProduct | NutritionProduct) | The primary subject of the genomic study |
  | Σ | 0..1 | Reference(Encounter) | The healthcare event with which this genomics study is associated |
  | 0..1 | dateTime | When the genomic study was started | |
  | 0..* | Reference(ServiceRequest | Task) | Event resources that the genomic study is based on | |
  | 0..1 | Reference(Practitioner | PractitionerRole) | Healthcare professional who requested or referred the genomic study | |
  | 0..* | Reference(Practitioner | PractitionerRole) | Healthcare professionals who interpreted the genomic study | |
  | 0..* | CodeableReference(Condition | Observation) | Why the genomic study was performed | |
  | 0..1 | canonical(PlanDefinition) | The defined protocol that describes the study | |
  | 0..1 | uri | The URL pointing to an externally maintained protocol that describes the study | |
  | 0..* | Annotation | Comments related to the genomic study | |
  | 0..1 | markdown | Description of the genomic study | |
  | 0..* | BackboneElement | Genomic Analysis Event | |
   | Σ | 0..* | Identifier | Identifiers for the analysis event |
   | Σ | 0..* | CodeableConcept | Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI) Binding: Genomic Study Method Type (Example) |
   | 0..* | CodeableConcept | Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change) Binding: Genomic Study Change Type (Example) | |
   | 0..1 | CodeableConcept | Genome build that is used in this analysis Binding: HumanRefSeqNCBIBuildId  (Extensible) (Extensible) | |
   | 0..1 | canonical(PlanDefinition | ActivityDefinition) | The defined protocol that describes the analysis | |
   | 0..1 | uri | The URL pointing to an externally maintained protocol that describes the analysis | |
   | Σ | 0..1 | string | Name of the analysis event (human friendly) |
   | Σ | 0..* | Reference(Any) | What the genomic analysis is about, when it is not about the subject of record |
   | Σ | 0..* | Reference(Specimen) | The specimen used in the analysis event |
   | 0..1 | dateTime | The date of the analysis event | |
   | 0..* | Annotation | Any notes capture with the analysis event | |
   | 0..1 | Reference(Procedure | Task) | The protocol that was performed for the analysis event | |
   | 0..* | Reference(DocumentReference | Observation) | The genomic regions to be studied in the analysis (BED file) | |
   | 0..* | Reference(DocumentReference | Observation) | Genomic regions actually called in the analysis event (BED file) | |
   | 0..* | BackboneElement | Inputs for the analysis event | |
    | Σ | 0..1 | Reference(DocumentReference) | File containing input data |
    | 0..1 | CodeableConcept | Type of input data (e.g., BAM, CRAM, or FASTA) Binding: Genomic Study Data Format (Example) | |
    | 0..1 | The analysis event or other GenomicStudy that generated this input file | ||
     | Identifier | |||
     | Reference(GenomicStudy) | |||
   | 0..* | BackboneElement | Outputs for the analysis event | |
    | Σ | 0..1 | Reference(DocumentReference) | File containing output data |
    | Σ | 0..1 | CodeableConcept | Type of output data (e.g., VCF, MAF, or BAM) Binding: Genomic Study Data Format (Example) |
   | 0..* | BackboneElement | Performer for the analysis event | |
    | 0..1 | Reference(Practitioner | PractitionerRole | Organization | Device) | The organization, healthcare professional, or others who participated in performing this analysis | |
    | 0..1 | CodeableConcept | Role of the actor for this analysis | |
   | 0..* | BackboneElement | Devices used for the analysis (e.g., instruments, software), with settings and parameters | |
    | 0..1 | Reference(Device) | Device used for the analysis | |
    | 0..1 | CodeableConcept | Specific function for the device used for the analysis | |
 Documentation for this format Documentation for this format  | ||||
See the Extensions for this resource
XML Template
<GenomicStudy xmlns="http://hl7.org/fhir"><!-- from Resource: id, meta, implicitRules, and language --> <!-- from DomainResource: text, contained, extension, and modifierExtension --> <identifier><!-- 0..* Identifier Identifiers for this genomic study --></identifier> <status value="[code]"/><!-- 1..1 registered | available | cancelled | entered-in-error | unknown --> <type><!-- 0..* CodeableConcept The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling) --></type> <subject><!-- 1..1 Reference(BiologicallyDerivedProduct|Group|NutritionProduct| Patient|Substance) The primary subject of the genomic study --></subject> <encounter><!-- 0..1 Reference(Encounter) The healthcare event with which this genomics study is associated --></encounter> <startDate value="[dateTime]"/><!-- 0..1 When the genomic study was started --> <basedOn><!-- 0..* Reference(ServiceRequest|Task) Event resources that the genomic study is based on --></basedOn> <referrer><!-- 0..1 Reference(Practitioner|PractitionerRole) Healthcare professional who requested or referred the genomic study --></referrer> <interpreter><!-- 0..* Reference(Practitioner|PractitionerRole) Healthcare professionals who interpreted the genomic study --></interpreter> <reason><!-- 0..* CodeableReference(Condition|Observation) Why the genomic study was performed --></reason> <instantiatesCanonical><!-- 0..1 canonical(PlanDefinition) The defined protocol that describes the study --></instantiatesCanonical> <instantiatesUri value="[uri]"/><!-- 0..1 The URL pointing to an externally maintained protocol that describes the study --> <note><!-- 0..* Annotation Comments related to the genomic study --></note> <description value="[markdown]"/><!-- 0..1 Description of the genomic study --> <analysis> <!-- 0..* Genomic Analysis Event --> <identifier><!-- 0..* Identifier Identifiers for the analysis event --></identifier> <methodType><!-- 0..* CodeableConcept Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI) --></methodType> <changeType><!-- 0..* CodeableConcept Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change) --></changeType> <genomeBuild><!-- 0..1 CodeableConcept Genome build that is used in this analysis
--></genomeBuild> <instantiatesCanonical><!-- 0..1 canonical(ActivityDefinition|PlanDefinition) The defined protocol that describes the analysis --></instantiatesCanonical> <instantiatesUri value="[uri]"/><!-- 0..1 The URL pointing to an externally maintained protocol that describes the analysis --> <title value="[string]"/><!-- 0..1 Name of the analysis event (human friendly) --> <focus><!-- 0..* Reference(Any) What the genomic analysis is about, when it is not about the subject of record --></focus> <specimen><!-- 0..* Reference(Specimen) The specimen used in the analysis event --></specimen> <date value="[dateTime]"/><!-- 0..1 The date of the analysis event --> <note><!-- 0..* Annotation Any notes capture with the analysis event --></note> <protocolPerformed><!-- 0..1 Reference(Procedure|Task) The protocol that was performed for the analysis event --></protocolPerformed> <regionsStudied><!-- 0..* Reference(DocumentReference|Observation) The genomic regions to be studied in the analysis (BED file) --></regionsStudied> <regionsCalled><!-- 0..* Reference(DocumentReference|Observation) Genomic regions actually called in the analysis event (BED file) --></regionsCalled> <input> <!-- 0..* Inputs for the analysis event --> <file><!-- 0..1 Reference(DocumentReference) File containing input data --></file> <type><!-- 0..1 CodeableConcept Type of input data (e.g., BAM, CRAM, or FASTA) --></type> <generatedBy[x]><!-- 0..1 Identifier|Reference(GenomicStudy) The analysis event or other GenomicStudy that generated this input file --></generatedBy[x]> </input> <output> <!-- 0..* Outputs for the analysis event --> <file><!-- 0..1 Reference(DocumentReference) File containing output data --></file> <type><!-- 0..1 CodeableConcept Type of output data (e.g., VCF, MAF, or BAM) --></type> </output> <performer> <!-- 0..* Performer for the analysis event --> <actor><!-- 0..1 Reference(Device|Organization|Practitioner|PractitionerRole) The organization, healthcare professional, or others who participated in performing this analysis --></actor> <role><!-- 0..1 CodeableConcept Role of the actor for this analysis --></role> </performer> <device> <!-- 0..* Devices used for the analysis (e.g., instruments, software), with settings and parameters --> <device><!-- 0..1 Reference(Device) Device used for the analysis --></device> <function><!-- 0..1 CodeableConcept Specific function for the device used for the analysis --></function> </device> </analysis> </GenomicStudy>
JSON Template
{ "resourceType" : "GenomicStudy",
// from Resource: id, meta, implicitRules, and language
// from DomainResource: text, contained, extension, and modifierExtension
"identifier" : [{ Identifier }], // Identifiers for this genomic study
"status" : "<code>", // R! registered | available | cancelled | entered-in-error | unknown
"type" : [{ CodeableConcept }], // The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling)
"subject" : { Reference(BiologicallyDerivedProduct|Group|NutritionProduct|
Patient|Substance) }, // R! The primary subject of the genomic study
"encounter" : { Reference(Encounter) }, // The healthcare event with which this genomics study is associated
"startDate" : "<dateTime>", // When the genomic study was started
"basedOn" : [{ Reference(ServiceRequest|Task) }], // Event resources that the genomic study is based on
"referrer" : { Reference(Practitioner|PractitionerRole) }, // Healthcare professional who requested or referred the genomic study
"interpreter" : [{ Reference(Practitioner|PractitionerRole) }], // Healthcare professionals who interpreted the genomic study
"reason" : [{ CodeableReference(Condition|Observation) }], // Why the genomic study was performed
"instantiatesCanonical" : "<canonical(PlanDefinition)>", // The defined protocol that describes the study
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the study
"note" : [{ Annotation }], // Comments related to the genomic study
"description" : "<markdown>", // Description of the genomic study
"analysis" : [{ // Genomic Analysis Event
"identifier" : [{ Identifier }], // Identifiers for the analysis event
"methodType" : [{ CodeableConcept }], // Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI)
"changeType" : [{ CodeableConcept }], // Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change)
"genomeBuild" : { CodeableConcept }, // Genome build that is used in this analysis
"resourceType" : "GenomicStudy",
// from Resource: id, meta, implicitRules, and language
// from DomainResource: text, contained, extension, and modifierExtension
"identifier" : [{ Identifier }], // Identifiers for this genomic study
"status" : "<code>", // R! registered | available | cancelled | entered-in-error | unknown
"type" : [{ CodeableConcept }], // The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling)
"subject" : { Reference(BiologicallyDerivedProduct|Group|NutritionProduct|
Patient|Substance) }, // R! The primary subject of the genomic study
"encounter" : { Reference(Encounter) }, // The healthcare event with which this genomics study is associated
"startDate" : "<dateTime>", // When the genomic study was started
"basedOn" : [{ Reference(ServiceRequest|Task) }], // Event resources that the genomic study is based on
"referrer" : { Reference(Practitioner|PractitionerRole) }, // Healthcare professional who requested or referred the genomic study
"interpreter" : [{ Reference(Practitioner|PractitionerRole) }], // Healthcare professionals who interpreted the genomic study
"reason" : [{ CodeableReference(Condition|Observation) }], // Why the genomic study was performed
"instantiatesCanonical" : "<canonical(PlanDefinition)>", // The defined protocol that describes the study
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the study
"note" : [{ Annotation }], // Comments related to the genomic study
"description" : "<markdown>", // Description of the genomic study
"analysis" : [{ // Genomic Analysis Event
"identifier" : [{ Identifier }], // Identifiers for the analysis event
"methodType" : [{ CodeableConcept }], // Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI)
"changeType" : [{ CodeableConcept }], // Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change)
"genomeBuild" : { CodeableConcept }, // Genome build that is used in this analysis  "instantiatesCanonical" : "<canonical(PlanDefinition|ActivityDefinition)>", // The defined protocol that describes the analysis
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the analysis
"title" : "<string>", // Name of the analysis event (human friendly)
"focus" : [{ Reference(Any) }], // What the genomic analysis is about, when it is not about the subject of record
"specimen" : [{ Reference(Specimen) }], // The specimen used in the analysis event
"date" : "<dateTime>", // The date of the analysis event
"note" : [{ Annotation }], // Any notes capture with the analysis event
"protocolPerformed" : { Reference(Procedure|Task) }, // The protocol that was performed for the analysis event
"regionsStudied" : [{ Reference(DocumentReference|Observation) }], // The genomic regions to be studied in the analysis (BED file)
"regionsCalled" : [{ Reference(DocumentReference|Observation) }], // Genomic regions actually called in the analysis event (BED file)
"input" : [{ // Inputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing input data
"type" : { CodeableConcept }, // Type of input data (e.g., BAM, CRAM, or FASTA)
// generatedBy[x]: The analysis event or other GenomicStudy that generated this input file. One of these 2:
"generatedByIdentifier" : { Identifier },
"generatedByReference" : { Reference(GenomicStudy) }
}],
"output" : [{ // Outputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing output data
"type" : { CodeableConcept } // Type of output data (e.g., VCF, MAF, or BAM)
}],
"performer" : [{ // Performer for the analysis event
"actor" : { Reference(Device|Organization|Practitioner|PractitionerRole) }, // The organization, healthcare professional, or others who participated in performing this analysis
"role" : { CodeableConcept } // Role of the actor for this analysis
}],
"device" : [{ // Devices used for the analysis (e.g., instruments, software), with settings and parameters
"device" : { Reference(Device) }, // Device used for the analysis
"function" : { CodeableConcept } // Specific function for the device used for the analysis
}]
}]
}
"instantiatesCanonical" : "<canonical(PlanDefinition|ActivityDefinition)>", // The defined protocol that describes the analysis
"instantiatesUri" : "<uri>", // The URL pointing to an externally maintained protocol that describes the analysis
"title" : "<string>", // Name of the analysis event (human friendly)
"focus" : [{ Reference(Any) }], // What the genomic analysis is about, when it is not about the subject of record
"specimen" : [{ Reference(Specimen) }], // The specimen used in the analysis event
"date" : "<dateTime>", // The date of the analysis event
"note" : [{ Annotation }], // Any notes capture with the analysis event
"protocolPerformed" : { Reference(Procedure|Task) }, // The protocol that was performed for the analysis event
"regionsStudied" : [{ Reference(DocumentReference|Observation) }], // The genomic regions to be studied in the analysis (BED file)
"regionsCalled" : [{ Reference(DocumentReference|Observation) }], // Genomic regions actually called in the analysis event (BED file)
"input" : [{ // Inputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing input data
"type" : { CodeableConcept }, // Type of input data (e.g., BAM, CRAM, or FASTA)
// generatedBy[x]: The analysis event or other GenomicStudy that generated this input file. One of these 2:
"generatedByIdentifier" : { Identifier },
"generatedByReference" : { Reference(GenomicStudy) }
}],
"output" : [{ // Outputs for the analysis event
"file" : { Reference(DocumentReference) }, // File containing output data
"type" : { CodeableConcept } // Type of output data (e.g., VCF, MAF, or BAM)
}],
"performer" : [{ // Performer for the analysis event
"actor" : { Reference(Device|Organization|Practitioner|PractitionerRole) }, // The organization, healthcare professional, or others who participated in performing this analysis
"role" : { CodeableConcept } // Role of the actor for this analysis
}],
"device" : [{ // Devices used for the analysis (e.g., instruments, software), with settings and parameters
"device" : { Reference(Device) }, // Device used for the analysis
"function" : { CodeableConcept } // Specific function for the device used for the analysis
}]
}]
}
Turtle Template
@prefix fhir: <http://hl7.org/fhir/> .[ a fhir:GenomicStudy; fhir:nodeRole fhir:treeRoot; # if this is the parser root # from Resource: .id, .meta, .implicitRules, and .language # from DomainResource: .text, .contained, .extension, and .modifierExtension fhir:identifier ( [ Identifier ] ... ) ; # 0..* Identifiers for this genomic study fhir:status [ code ] ; # 1..1 registered | available | cancelled | entered-in-error | unknown fhir:type ( [ CodeableConcept ] ... ) ; # 0..* The type of the study (e.g., Familial variant segregation, Functional variation detection, or Gene expression profiling) fhir:subject [ Reference(BiologicallyDerivedProduct|Group|NutritionProduct|Patient|Substance) ] ; # 1..1 The primary subject of the genomic study fhir:encounter [ Reference(Encounter) ] ; # 0..1 The healthcare event with which this genomics study is associated fhir:startDate [ dateTime ] ; # 0..1 When the genomic study was started fhir:basedOn ( [ Reference(ServiceRequest|Task) ] ... ) ; # 0..* Event resources that the genomic study is based on fhir:referrer [ Reference(Practitioner|PractitionerRole) ] ; # 0..1 Healthcare professional who requested or referred the genomic study fhir:interpreter ( [ Reference(Practitioner|PractitionerRole) ] ... ) ; # 0..* Healthcare professionals who interpreted the genomic study fhir:reason ( [ CodeableReference(Condition|Observation) ] ... ) ; # 0..* Why the genomic study was performed fhir:instantiatesCanonical [ canonical(PlanDefinition) ] ; # 0..1 The defined protocol that describes the study fhir:instantiatesUri [ uri ] ; # 0..1 The URL pointing to an externally maintained protocol that describes the study fhir:note ( [ Annotation ] ... ) ; # 0..* Comments related to the genomic study fhir:description [ markdown ] ; # 0..1 Description of the genomic study fhir:analysis ( [ # 0..* Genomic Analysis Event fhir:identifier ( [ Identifier ] ... ) ; # 0..* Identifiers for the analysis event fhir:methodType ( [ CodeableConcept ] ... ) ; # 0..* Type of the methods used in the analysis (e.g., FISH, Karyotyping, MSI) fhir:changeType ( [ CodeableConcept ] ... ) ; # 0..* Type of the genomic changes studied in the analysis (e.g., DNA, RNA, or AA change) fhir:genomeBuild [ CodeableConcept ] ; # 0..1 Genome build that is used in this analysis fhir:instantiatesCanonical [ canonical(ActivityDefinition|PlanDefinition) ] ; # 0..1 The defined protocol that describes the analysis fhir:instantiatesUri [ uri ] ; # 0..1 The URL pointing to an externally maintained protocol that describes the analysis fhir:title [ string ] ; # 0..1 Name of the analysis event (human friendly) fhir:focus ( [ Reference(Any) ] ... ) ; # 0..* What the genomic analysis is about, when it is not about the subject of record fhir:specimen ( [ Reference(Specimen) ] ... ) ; # 0..* The specimen used in the analysis event fhir:date [ dateTime ] ; # 0..1 The date of the analysis event fhir:note ( [ Annotation ] ... ) ; # 0..* Any notes capture with the analysis event fhir:protocolPerformed [ Reference(Procedure|Task) ] ; # 0..1 The protocol that was performed for the analysis event fhir:regionsStudied ( [ Reference(DocumentReference|Observation) ] ... ) ; # 0..* The genomic regions to be studied in the analysis (BED file) fhir:regionsCalled ( [ Reference(DocumentReference|Observation) ] ... ) ; # 0..* Genomic regions actually called in the analysis event (BED file) fhir:input ( [ # 0..* Inputs for the analysis event fhir:file [ Reference(DocumentReference) ] ; # 0..1 File containing input data fhir:type [ CodeableConcept ] ; # 0..1 Type of input data (e.g., BAM, CRAM, or FASTA) # generatedBy[x] : 0..1 The analysis event or other GenomicStudy that generated this input file. One of these 2 fhir:generatedBy [ a fhir:Identifier ; Identifier ] fhir:generatedBy [ a fhir:Reference ; Reference(GenomicStudy) ] ] ... ) ; fhir:output ( [ # 0..* Outputs for the analysis event fhir:file [ Reference(DocumentReference) ] ; # 0..1 File containing output data fhir:type [ CodeableConcept ] ; # 0..1 Type of output data (e.g., VCF, MAF, or BAM) ] ... ) ; fhir:performer ( [ # 0..* Performer for the analysis event fhir:actor [ Reference(Device|Organization|Practitioner|PractitionerRole) ] ; # 0..1 The organization, healthcare professional, or others who participated in performing this analysis fhir:role [ CodeableConcept ] ; # 0..1 Role of the actor for this analysis ] ... ) ; fhir:device ( [ # 0..* Devices used for the analysis (e.g., instruments, software), with settings and parameters fhir:device [ Reference(Device) ] ; # 0..1 Device used for the analysis fhir:function [ CodeableConcept ] ; # 0..1 Specific function for the device used for the analysis ] ... ) ; ] ... ) ; ]
Additional definitions: Master Definition XML + JSON, XML Schema/Schematron + JSON Schema, ShEx (for Turtle) + see the extensions, the spreadsheet version & the dependency analysis
| Path | ValueSet | Type | Documentation |
|---|---|---|---|
| GenomicStudy.status | GenomicStudyStatus | Required | The status of the GenomicStudy. |
| GenomicStudy.type | GenomicStudyType | Example | The type of the GenomicStudy. |
| GenomicStudy.analysis.methodType | GenomicStudyMethodType | Example | The method type of the GenomicStudy analysis. |
| GenomicStudy.analysis.changeType | GenomicStudyChangeType | Example | The change type of the GenomicStudy analysis. |
| GenomicStudy.analysis.genomeBuild | http://loinc.org/vs/LL1040-6  | Extensible | |
| GenomicStudy.analysis.input.type | GenomicStudyDataFormat | Example | The data formats relevant to Genomic Study analysis. |
| GenomicStudy.analysis.output.type | GenomicStudyDataFormat | Example | The data formats relevant to Genomic Study analysis. |
GenomicStudy uses many example codes for terminology binding. Reviewers and implementers are strongly encouraged to provide their comments and feedback about the example codes for terminology binding purposes.
Below the attributes of provided example bindings are listed. Links are included to indicate where the example codes from were pulled from.
The type example codes were based on discussions by Clinical Genomics Workgroup.
The method type example codes were pulled from National Library of Medicine-Genetic Testing Registry (NCBI-GTR)  values of describing different testing methods on various levels: major method category
values of describing different testing methods on various levels: major method category  , method category
, method category  , and primary methodology
, and primary methodology  .
.
The input and output type example codes were pulled from Integrative Genomics Viewer Documentation  by Broad Institute.
by Broad Institute.
The change type example codes were based on discussions by Clinical Genomics Workgroup.
GenomicStudy.description is a markdown datatype, and implementers should carefully consider how to appropriately handle this attribute. The characters in markdown formatting can conflict with those commonly used in descriptions of genomic data. In particular, descriptions that contain mentions of "star alleles" (widely used in the pharmacogenomics and HLA domains) could be munged in a way that prevents accurate interpretation. For example, consider this text:
This genomic study analyzes CYP2D6*1 and CYP2D6*2
If the system producing this data treats this as a simple text string with no special processing, but a receiving system processes this via a markdown rendering engine, the two '*' characters would be processed as markdown formatting characters. This would italize the text between '*' characters, and not display '*' characters. This could cause an inaccurate interpretation of the study description.
There are several ways data producers can ensure content is appropriately rendered by receiving systems without requiring the content to be formatted as markdown prior to sending. Here are three basic approaches to consider:
Escaping individual characters (with a \) that act as markdown formatting characters:
This genomic study analyzes CYP2D6\*1 and CYP2D6\*2
Escape words (with a `) that contain markdown formatting characters:
This genomic study analyzes `CYP2D6*1` and `CYP2D6*2`
Escape full text blocks (with a ```) that contain markdown formatting characters:
```This genomic study analyzes CYP2D6*1 and CYP2D6*2```
GenomicStudy.subject and GenomicStudy.analysis.focus can reference many resource types besides Patient such as Group, BiologicallyDerivedProduct, or Substance. In addition, it can provide more details about involved genomic files as inputs or outputs. These various options allow the GenomicStudy resource to cover many use cases besides direct patient care, e.g., research studies that involve multiple patients or environmental samples. Through the following subsections, some of these use cases are described.
Trio studies involve a proband and two more subjects such as proband’s mother and father for a de novo mutation detection study. GenomicStudy would list the proband as GenomicStudy.subject because it is the main subject of the study. Each of the study participants, i.e., proband, mother, and father, may have their own GenomicStudy.analysis entry. When an analysis was performed on an entity other than the GenomicStudy.subject, the GenomicStudy.analysis.focus attribute would reference that entity. If a GenomicStudy.analysis entry documented the analysis of all participants of the trio, each participant would be referenced by GenomicStudy.analysis.focus.
GenomicStudy.analysis.input lists various files that may be used for each individual analysis, their types, and their generation context. GenomicStudy.analysis.input.file elements may link these files to DocumentReference resources that provides more details about each individual files. One of the main details is the subject of a file. If a file is linked to a specific patient, the corresponding DocumentReference.subject may reference this Patient Resource. If the file contains data from multiple persons, the corresponding DocumentReference.subject may reference a Group resource that lists these persons, and their relationship to each other if available.
Somatic mutation studies may use multiple samples from the patient to support mutation detection, e.g., tumor-normal sample pair. GenomicStudy.analysis may describe conducted analyses per each sample, where GenomicStudy.analysis.specimen elements may provide more details about each individual element. In addition GenomicStudy.analysis.input can list relevant input files, where the DocumentReference referenced by GenomicStudy.analysis.input.file can list the specific specimen this input file is related to using DocumentReference.subject as a Specimen resource instance.
Search parameters for this resource. See also the full list of search parameters for this resource, and check the Extensions registry for search parameters on extensions related to this resource. The common parameters also apply. See Searching for more information about searching in REST, messaging, and services.
| Name | Type | Description | Expression | In Common |
| focus | reference | What the genomic study analysis is about, when it is not about the subject of record | GenomicStudy.analysis.focus (Any) | |
| identifier | token | Identifiers for the Study | GenomicStudy.identifier | |
| patient | reference | Who the study is about | GenomicStudy.subject.where(resolve() is Patient) (Group, BiologicallyDerivedProduct, NutritionProduct, Patient, Substance) | |
| status | token | The status of the study | GenomicStudy.status | |
| subject | reference | Who the study is about | GenomicStudy.subject (Group, BiologicallyDerivedProduct, NutritionProduct, Patient, Substance) |