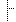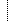| Error | StructureDefinition.version | Values for version differ: '1.0.0' vs '1.1.0' |
| Information | StructureDefinition.status | Values for status differ: 'draft' vs 'active' |
| Information | StructureDefinition.date | Values for date differ: '2017-05-20T00:00:00-04:00' vs '2021-04-13T19:13:37+00:00' |
| Information | StructureDefinition.publisher | Values for publisher differ: 'HL7 Clinical Genomics WG' vs 'HL7 International Clinical Genomics Work Group' |
| Information | StructureDefinition.jurisdiction | Added the item 'http://unstats.un.org/unsd/methods/m49/m49.htm#001' |
| Warning | Observation | Elements differ in short:
'Haplotype'
'Measurements and simple assertions' |
| Warning | Observation | Elements differ in definition:
'A finding that a patient has a specific haplotype.'
'Measurements and simple assertions made about a patient, device or other subject.' |
| Warning | Observation | Elements differ in comments:
'Defines those properties that are common for all findings of genetic characteristics - namely genotype, haplotype and any of the types of variants.'
'Used for simple observations such as device measurements, laboratory atomic results, vital signs, height, weight, smoking status, comments, etc. Other resources are used to provide context for observations such as laboratory reports, etc.' |
| Warning | Observation.status | Elements differ in comments:
'… This should generally be 'completed' or 'revised'.'
'This element is labeled as a modifier because the status contains codes that mark the resource as not currently valid.' |
| Warning | Observation.category | Elements differ in comments:
'… This should be populated with 'lab'.'
'In addition to the required category valueset, this element allows various categorization schemes based on the owner’s definition of the category and effectively multiple categories can be used at once. The level of granularity is defined by the category concepts in the value set.' |
| Warning | Observation.subject | Elements differ in comments:
'… This must be the same patient as on the DiagnosticReport unless combining data from multiple fetuses or family members in a single report.'
'One would expect this element to be a cardinality of 1..1. The only circumstance in which the subject can be missing is when the observation is made by a device that does not know the patient. In this case, the observation SHALL be matched to a patient through some context/channel matching technique, and at this point, the observation should be updated.' |
| Warning | Observation.method | Elements differ in short:
'Haplotype determination method'
'How it was done' |
| Warning | Observation.method | Elements differ in definition:
'Indicates the method used to determine the haplotype.'
'Indicates the mechanism used to perform the observation.' |
| Warning | Observation.specimen | Elements differ in short:
'Specimen examined'
'Specimen used for this observation' |
| Warning | Observation.specimen | Elements differ in definition:
'The specimen examined to determine genetic information.'
'The specimen that was used when this observation was made.' |
| Warning | Observation.derivedFrom | Elements differ in definition:
'variant or haplotype from which this haplotype is derived.'
'The target resource that represents a measurement from which this observation value is derived. For example, a calculated anion gap or a fetal measurement based on an ultrasound image.' |
 abstract
abstract baseDefinition
baseDefinition copyright
copyright date
date description
description experimental
experimental fhirVersion
fhirVersion jurisdiction
jurisdiction
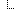 jurisdiction[0]
jurisdiction[0] kind
kind name
name publisher
publisher purpose
purpose status
status title
title type
type url
url version
version