

This page is part of the HL7 FHIR Implementation Guide: minimal Common Oncology Data Elements (mCODE) Release 1 - US Realm | STU1 (v0.9.1: STU 1 Ballot 1) based on FHIR (HL7® FHIR® Standard) R4. The current version which supersedes this version is 4.0.0. For a full list of available versions, see the Directory of published versions
According to the National Cancer Institute, 38.5 percent of men and women will be diagnosed with cancer at some point during their lifetimes. In 2014, an estimated 14.7M people were living with cancer in the United States. While these numbers are staggering, the silver lining in the wide prevalence of cancer is the potential to learn from treatment of millions of patients. If we had research-quality data from all cancer patients, it would enable higher quality health outcomes. Today, we lack the data models, technologies, and methods to capture that data.
mCODE™ (short for Minimal Common Oncology Data Elements) is an initiative intended to assemble a core set of structured data elements for oncology electronic health records (EHRs). mCODE™ is a step towards capturing research-quality data from the treatment of all cancer patients. This would enable the treatment of every cancer patient to contribute to comparative effectiveness analysis (CEA) of cancer treatments. mCODE™ has been created and is being supported by the American Society of Clinical Oncology (ASCO®)in collaboration with the MITRE Corporation.
In late 2018, ASCO convened committee of twenty leading clinical experts in oncology, radiology, surgery, and public health developed two use cases that drove the initial clinical data requirements for mCODE:
While mCODE ultimately is meant to be applicable to across all types of cancer, the initial focus (and both use cases) has been on solid tumors.
In addition to information obtained from subject matter experts, several pre-existing standards, nomenclatures, and guidelines were consulted in the development of this specification, including:
After initial development, in early 2019, an open survey was conducted to validate and prioritize the data elements from these use cases. Further down-scoping was done based on whether the data would be stored or capture in an electronic health record (EHR), and if it would place undue documentation burden on clinicians.
The data elements identified in this process were modeled using the Clinical Information Modeling and Profiling Language (CIMPL) and exported as FHIR Profiles. The profiles, related FHIR artifacts, and other technical implementation information, constitute the bulk of this IG. What follows is an overview of mCODE, directed primarily at clinical readers. Readers should also take note of the Data Dictionary (Excel download), a simplified, flattened list of mCODE elements.
mCODE consists of data elements divided into six groups, illustrated in the following diagram:
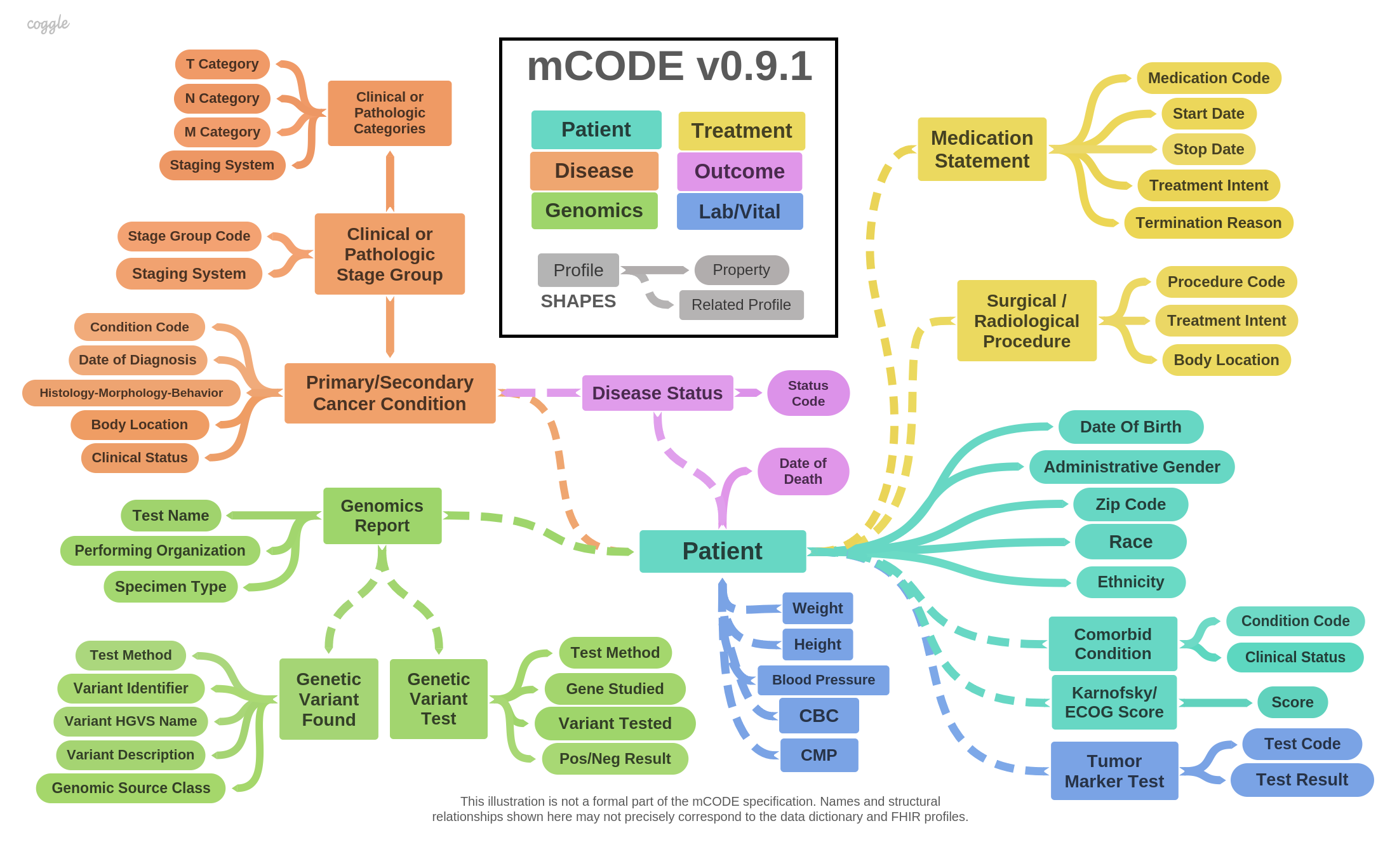
The mCODE Patient group contains the following basic information about the patient:
Patient is the most essential FHIR profile, as all other mCODE major elements reference it. The mCODE Patient profile differs only slightly from the US Core Patient Profile. Most significantly, Patient.deceased is a must-support element in mCODE.
The mCODE Disease Characterization group includes data elements specific to the diagnosis and staging of cancer. This includes:
The cancer diagnosis combines the type, site, and certain characteristics of the cancer. Depending on the EHR and provider organization, different code systems may be used, such as:
Because each of these coding systems are found "in the wild", mCODE supports all three. Implementers should be aware, however, that how the cancer diagnosis is coded can affect compliance with US Core (see Implementation Notes for details). Two attributes and one FHIR extension of the FHIR Condition Resource are involved with coding the cancer diagnosis: the Code, the HistologyMorphologyBehavior extension, and the Body Site. How these attributes are used, depending on the coding system, is captured in the table below:

Implementers should reference the PrimaryCancerCondition and Secondary Cancer Condition profiles for details on the use of these terminologies and associated value sets.
Cancer stage information is contained in a set of profiles, representing clinical stage group and pathologic stage group panels with members representing the primary tumor (T) category, the regional nodes (N) category, and the distant metastases (M) category. Non-TNM staging systems are not currently represented in mCODE, reflecting mCODE's current focus on solid tumors. In mCODE, a single patient may have more than one staging panel, although this is not common in practice.
Clinical applications vary in their representation of T, N, and M staging category values, falling into one of two naming conventions:
mCODE recommends that the implementer align with AJCC's convention of representing the staging category value with the prepended classification in both TNMClinicalStageGroup and TNMPathologicStageGroup profiles. This code convention is aligned with the AJCC's digital data and clearly distinguishes the staging classification as clinical, pathologic, or neoadjuvant without having to retrieve further context from the model. Nonetheless, separate profiles for clinical and pathological staging were developed, with an eye toward future extensibility, in particular, the ability to additional prognostic factors relevant to particular types of cancers in the TNMPathologicStageGroup.
Many laboratory tests could be relevant to an individual with cancer. The initial mCODE release includes only two core laboratory tests:
These core diagnostic labs are modeled separately from cancer-specific serum and tissue-based prognostic factors.
Tumor markers are key prognostic factors in calculating cancer staging, identifying treatment options, and monitoring progression of disease. For example, an abnormal increase in prostate-specific antigen (PSA) levels is a prognostic factor for prostate cancer. Other tumor markers include estrogen receptor (ER) status, progresterone receptor (PR) status, carcinoembryonic antigen (CEA) levels, among others. See the profile TumorMarkerTest for full details.
We distinguish Tumor Marker Tests from genetic tests that are measured at the DNA, RNA, or chromosomal level, addressed in the Genomics section.
Vital signs are measurements of the most essential, or "vital" body functions. Traditionally, vital signs include blood pressure, heart rate, respiratory rate, and temperature. More recently, height and weight have been included. Only BP, body height and body weight are included in mCode because they are believed to be the most critical to assessment and treatment.
The vital sign profiles defined by mCODE are consistent with the FHIR vital sign profiles, which are incorporated by reference into US Core v3. The difference between FHIR and mCODE vital signs is that mCODE provides for reporting of preconditions, body positions, blood pressure method, and blood pressure body location, with appropriate value sets. The vital signs model in mCODE is aligned with the Vital Signs Implementation Guide being developed in cooperation with the Clinical Information Modeling Initiative (CIMI) Work Group. Although mCODE defines its own vital signs profiles, if and when detailed vital signs profiles are standardized in a widely-accepted FHIR IG, mCODE will likely switch over to those profiles.
The Treatment group includes reporting of procedures and medications used to treat a cancer patient, or relevant to that treatment. Treatments are captured using the following profiles:
Like US Core, mCODE gives preference to representing medications using the National Library of Medicine (NLM) RxNorm terminology - a coding standard established by the Office of the National Coordinator (ONC) for the exchange of drugs. However, RxNorm is restricted to FDA-approved drugs and does not include clinical trial drugs. To address this limitation, mCODE allows for the inclusion of other coding systems like the NCI Thesaurus (NCIT) to represent clinical trial oncology drugs.
mCODE includes the minimal set of genetic related elements relevant to capture in an EHR to inform cancer assessment and treatment options. The approach is based on the HL7 CGWG Clinical Genomics Reporting Implementation Guide. However, mCODE simplifies genomics reporting to single discrete variants or to variants that were found in a given DNA region. Three profiles relate to the capture of clinical genomics data:
The identity of non-genomic laboratory tests is typically represented by a Logical Observation Identifiers and Names (LOINC) code. However, many genetic tests and panels do not have LOINC codes, although some might have an identifier in the NCBI Genetic Testing Registry (GTR), a central location for voluntary submission of genetic test information by providers. While GTR is currently the best source for identifying many genetic tests, the user should be aware that the GTR may not be reliable source since the test data is voluntarily updated and there is no overarching data steward. Standardization of codes for genetic tests is essential to facilitate data analysis of genetic tests, and should be a priority for the genomics testing community in the near future. Implementers should also note that, to conform to the requirements of the US Core Laboratory Result Profile, LOINC must be used, if a suitable code is available. If there is no suitable code in LOINC, then a code from an alternative code system (such as GTR) can be used.
Recording outcomes of cancer treatment in mCODE involves two data elements: disease status and date of death. Other common outcome measures, such as progression-free survival, time to recurrence, and overall survival, can be derived from time-indexed observations of disease status. The date of diagnosis is also required for some derived measures (see Disease Characterization). At this time, mCODE does not include patient reported outcomes.
Formal recording of disease status is often limited to clinical trials, involving precise criteria such as RECIST. The lack of outcome data outside of trials greatly limits the application of real-world data. Disease status information is rarely found in structured form in EHRs. If recorded at all, the information is found in clinical notes, which is of limited usefulness.
mCODE asks for disease progression to be recorded in structured form as part of patient encounters. In mCODE, disease status is defined as "A clinician's qualitative judgment on the current trend of the cancer, e.g., whether it is stable, worsening (progressing), or improving (responding). The judgment may be based a single type or multiple kinds of evidence, such as imaging data, assessment of symptoms, tumor markers, laboratory data, etc." In other words, the disease status is an assessment by the oncologist that synthesizes all currently available information about the patient. The ICAREdata™ Project is conducting a study in association with a randomized controlled trial (RCT), which aims to demonstrate the ability to calculate equivalent clinical trial endpoints using computable clinical treatment data.
Date of death data can be obtained from several sources outside of the clinical setting. If available in the EHR, it can be reported through via mCODE, but more likely, it will be filled in from vital records, after the last clinical interaction.
The authors recognize the leadership and sponsorship of Dr. Monica Bertagnolli, President, ASCO and Dr. Jay Schnitzer, MITRE Chief Technology Officer. Dr. Steven Piantadosi and the Alliance for Clinical Trials in Oncology coordinated real-world data collection in clinical trials, as part of this project. The ASCO/CancerLinQ team was led by Dr. Robert Miller and Dr. Wendy Rubinstein. Lead MITRE contributors were Mark Kramer, Rute Martins, Chris Moesel, and May Terry. Andre Quina and Dr. Brian Anderson guide the overall mCODE effort at MITRE. HL7 sponsorship and input from Clinical Interoperability Council and Clinical Information Modeling Initiative is gratefully acknowledged, with special thanks to Richard Esmond and Laura Heermann Langford.
This IG was authored by the MITRE Corporation using the Clinical Information Modeling and Profiling Language (CIMPL), a free, open source toolchain from MITRE Corporation.
|
General Inquiries: |
|
|
Co-Editor: |
Dr. Robert Miller ASCO CancerLinQ |
|
Co-Editor: |
Mark Kramer MITRE Corporation |